To the editor
Huang et al.1 recently reported a crystal structure of a thermostabilised avian β1-adrenergic receptor (β1AR) that was described as being in a ligand-free basal state. On inspection of the electron density maps provided by the electron density server (eds.bmc.uu.se) for the corresponding PDB entry (4GPO) we observed significant unmodelled electron density in the ligand binding pocket. In view of this unexpected result, an unusual distribution of atomic temperature factors in the deposited coordinates and the very high affinity of cyanopindolol for β1AR that was used in the receptor preparation, we have repeated the structure determination using the deposited structure factors.
The structure was solved by molecular replacement with PHASER2 using the structure of cyanopindolol-bound thermostabilised avian β1AR-m233 (chain B of PDB entry 2VT4) following removal of all ligands, detergent molecules and waters. This model was used in preference to the structure of the salbutamol-bound form (PDB entry 2Y04) chosen by Huang et al because it was determined at higher resolution (2.7 Å for 2VT4 vs 3.05 Å for 2Y04). Chain B was selected because it is the most complete model and does not display the kinked helix 1 found in chains A and C. PHASER produced a clear solution with two molecules in the crystallographic asymmetric unit, and this model was refined with REFMAC54 applying automatic NCS restraints and restraining the model to a high resolution (2.1 Å) structure of β1AR-m23 (PDB entry 4BVN) with PROSMART5. The resulting electron density map displayed good density for cyanopindolol in both subunits, although it was stronger in chain A (Figure 1). Cyanopindolol was added to the model and another round of refinement carried out without the PROSMART restraints but now using the jelly-body restraints in REFMAC5. In this refinement, the cyanopindolol ligand was assigned an initial occupancy of 0.8 in both subunits and this was refined. The resulting model gave Rwork and Rfree values of 34.4% and 38.2%, with bond length r.m.s.d. value of 0.013 Å and the cyanopindolol occupancies refined to 1.00 for chain A and 0.84 for B. The electron density was well defined for the ligand in both chains (Figure 1). Fixing the occupancy of cyanopindolol to zero and repeating this refinement gave significantly higher Rwork and Rfree values of 35.2% and 38.9%.
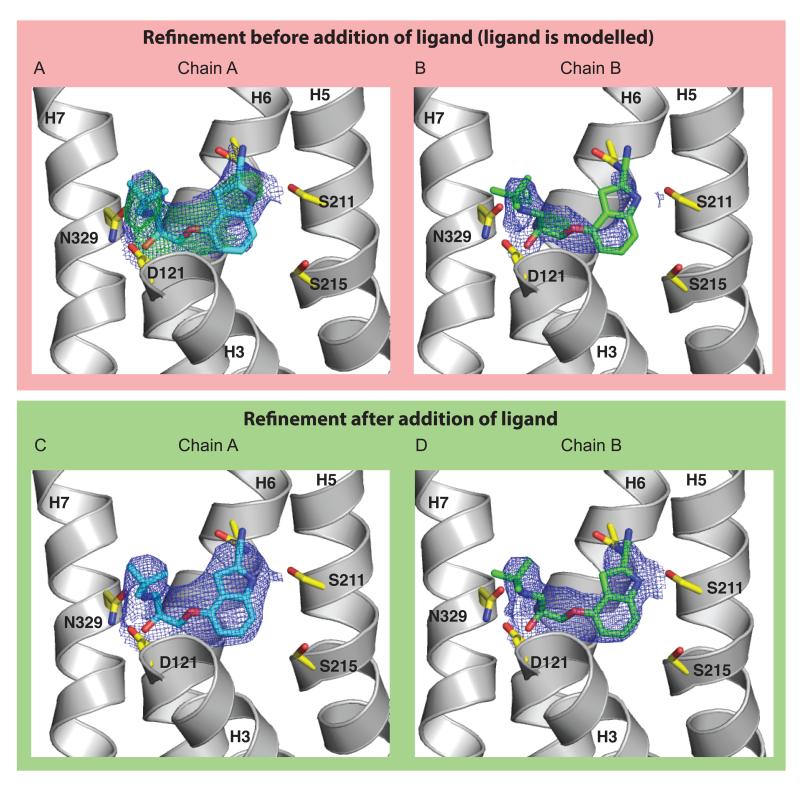
Figure 1.
Electron density for the cyanopindolol ligand. 2Fo-Fc electron density maps within 2 Å of cyanopindolol are shown for chains A and B. The transmembrane helices H3, H5, H6 and H7 are labeled and side chains forming the ligand binding pocket are indicated. Helices 1, 2, 4 and the upper part of helix 3 have been removed for clarity. (a, b) Following molecular replacement and one round of refinement with no ligand included in the model (the ligand is shown in its position in the final model). Density for the 2Fo-Fc maps (blue) is contoured at 1.3 σ (a) or 1.0 σ (b), with the Fo-Fc difference map (green) contoured at 3.0 σ. (c, d) Final density after inclusion of cyanopindolol in the model, with refined occupancies of 1.00 and 0.84 in chains A and B respectively. Density for the 2Fo-Fc maps (blue) is contoured at 2.0 σ (c) or 1.3 σ (d).
The quality of the ligand density (especially before its inclusion in the model) together with the high values for the refined occupancies and the increase in refinement R-factors when the ligand occupancy is set to zero provide convincing evidence that cyandopindolol is bound in the crystal and that the structure does not represent a ligand free state. This is consistent with the very high affinity of cyanopindolol for β1AR (13 pM)6 and, as Huang and colleagues found, it is very difficult to remove by dialysis due to its extremely slow off-rate. Only an experiment using radiolabelled cyanopindolol bound to purified β1AR to monitor directly ligand removal by dialysis, performed on the sample used to create the crystals, could provide convincing evidence that all the cyanopindolol was actually removed and was therefore absent in any crystals produced.
The R-factors we report are significantly higher than those reported by Huang et al. (Rwork 30.99%, Rfree 35.46%). There are two possible explanations for this. The first is differences in modeling the solvent contribution in PHENIX7 and REFMAC5. The second is the unusual distribution of atomic B-factors in the 4GPO coordinate file, where there are large differences in B-factor for bonded side chain atoms, with B factors both increasing and decreasing along the length of the side chain (eg Met36 in chain A has B factors of 75, 30, 24, 118, 128 Å2 for CA, CB, CG, SD, CE atoms respectively). This variation does not seem physically reasonable and may represent over-fitting of the data. It would also have been useful to carry out refinement against the original data, rather than the anisotropically truncated and sharpened data that were deposited, but this was not made available to us. The R-factors for our final model are within the expected range for a structure refined at this resolution.
A consequence of the presence of cyanopindolol in the 4GPO crystal structure is that the dimensions of the ligand binding pocket needs to be reassessed. Specifically, the contraction of the ligand binding pocket reported by Huang et al is no longer feasible due to steric clashes between the ligand and the side chains on helix 5. The ligand binding pocket in the structure we have refined from the same data exhibits identical dimensions to the original cyanopindolol-bound structure (2VT4).
The discrepancies we have described in the 4GPO structure are in the ligand binding pocket. The interesting packing of β1AR monomers within the crystal to form parallel dimers, which could be of physiological relevance, is unaffected upon re-refining the data.
Supplementary Material
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
The coordinate and reflection files (MTZ format) of the refined model area are available from the files beta1_final.pdb and beta1_final.mtz, respectively.
REFERENCES
- 1.Huang J, Chen S, Zhang JJ, Huang X-Y. Nat. Struct. Mol. Biol. 2013;20:419–425. doi: 10.1038/nsmb.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCoy AJ, et al. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warne T, et al. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 5.Nicholls RA, Long F, Murshudov GN. Acta Crystallogr. D Biol. Crystallogr. 2012;68:404–417. doi: 10.1107/S090744491105606X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker JG. PLoS One. 5:e15487. doi: 10.1371/journal.pone.0015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams PD, et al. Acta Crystallogr D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.