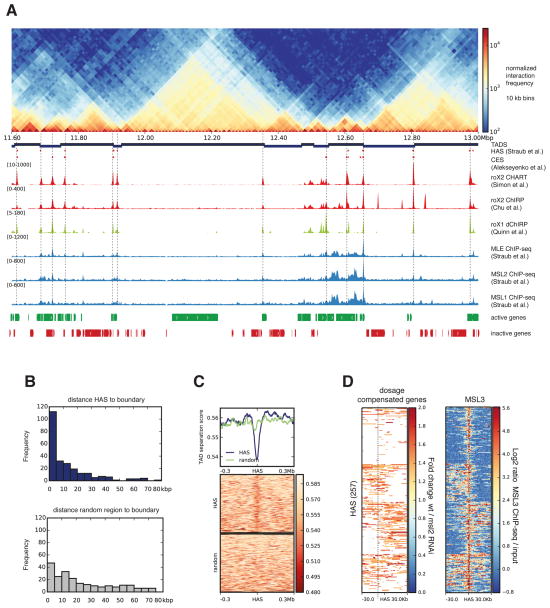
Figure 1. High affinity sites (HAS) are enriched at TAD boundaries.
A. Normalized Hi-C counts at 10 kb resolution for the region 11.6-13.0 Mb in chromosome X of S2 cells. From top, the tracks are as follows: partitioning of the genome into topologically associating domains (TADs); HAS as defined by Straub et al. (Straub et al., 2008); HAS reported by (Alekseyenko et al., 2008) (originally called chromosome entry sites (CES)); roX2 CHART (Simon et al., 2011); roX2 ChIRP (Chu et al., 2011); roX1 dChIRP (Quinn et al., 2014); MLE-, MSL1-, MSL2- ChIP-seq (Straub et al., 2013); active and inactive genes in S2 cells (Cherbas et al., 2011). Vertical lines are high resolution HAS based on roX2 and MSL2 binding. B. Distribution of distances from the boundaries to HAS (top, in blue) and from boundaries to the same number of shuffled random regions (bottom, in grey) within chromosome X. X-axis represents the distance in kb from HAS to boundary in bins of 5 kb, y-axis contains the number of HAS per bin. C. TAD separation score (see Extended Experimental Procedures) around HAS shows the tendency of the MSL complex to land at boundaries. Lower scores indicate better TAD separation. D. Dosage compensated genes (based on RNA-seq data from wild type and MSL2 RNAi treated S2 cells (Zhang et al., 2010)) and MSL3 ChIP-seq (Straub et al., 2013) up to 30 kb away from HAS. For the dosage compensation panel, only genes showing activity are colored. The heatmap rows were divided into four clusters based on the kmeans algorithm using deepTools (Ramírez et al., 2014). See also Table S1 and S2 and Figure S1, S2 and S3.