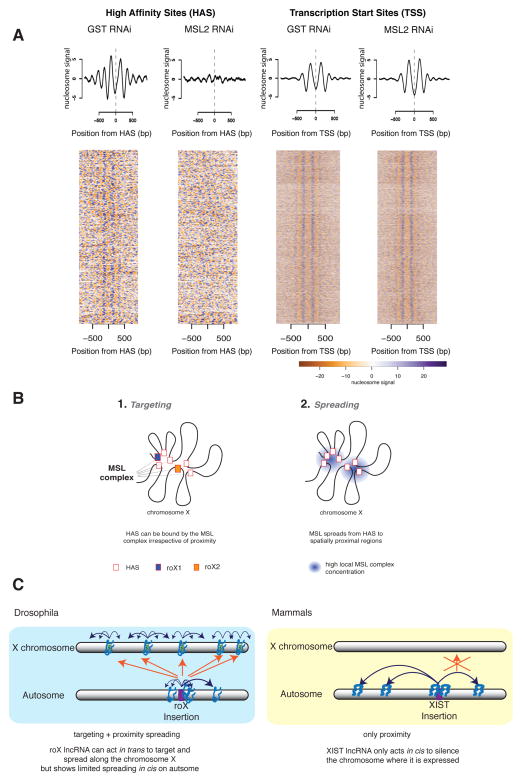
Figure 7. Depletion of MSL complex severely affects nucleosome positioning at HAS but not at TSS.
A. Summary plots (top) and heat maps (bottom) showing normalized nucleosome occupancy for regions centered around HAS (left, n= 257) and active TSS (right, n= 5985) in control (GST RNAi) and MSL2-depleted cells (MSL2 RNAi). Scale bar representing nucleosome signal is shown below. B. Conformation-based affinity model: we propose a model in which (1.) chromosome X is targeted via HAS by the MSL complex independently of spatial proximity. Then in (2.) the complex spreads from the HAS to spatially proximal regions. In (1.), the MSL complex binds specific regions of chromosome X containing a sequence motif, (the MRE), if this appears at the end of active genes that are at TAD boundaries. In (2.) the complex spreads (probably by diffusion) from HAS to spatially close regions. Because TAD boundaries appear enriched in contacts within each other as well as to other regions it is expected that they are physically close, thus offering an optimal location from which to reach all active genes on chromosome X. C. Comparison of ectopic expression of roX RNA versus Xist RNA on autosomes. Blue arrows denote spreading of the lncRNA in 3D proximity; orange arrows denote targeting of the lncRNA in trans. (Blue box) In Drosophila, ectopically expressed roX displays restricted spreading on the autosome indicating that spatial proximity is beneficial but not sufficient for roX RNA spreading. Importantly, roX RNA expressed from an autosomal location specifically targets the X in trans, leading to coating of the entire X chromosome by roX/MSL complex independent of spatial proximity. (Yellow box) In mammals, ectopically expressed Xist RNA can spread and coat the autosome using spatial proximity (Engreitz et al., 2013; Jiang et al., 2013; Simon et al., 2013) without targeting the X in trans. See also Figure S7.