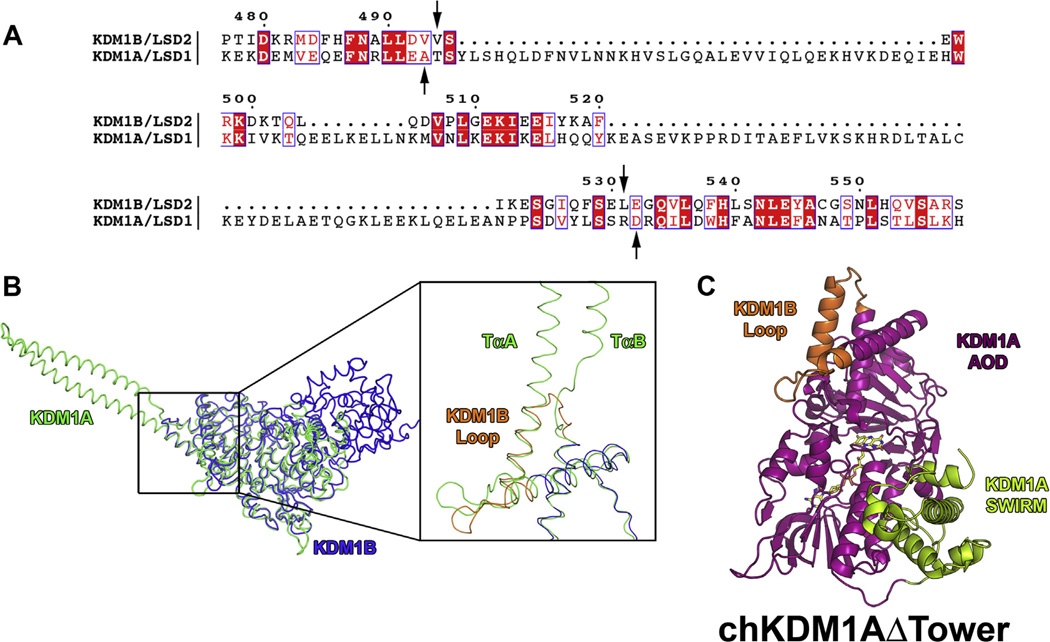
Fig. 2.
Sequence and structural alignment of KDM1A and KDM1B from H. sapiens and structural model of chKDM1AΔTower. Only residues 171–852 of KDM1A were used for alignment as per Karytinos et al. (A) Sequence alignment of KDM1A and KDM1B. Numbering is based on the primary amino acid sequence of KDM1B. Residues that are invariant in the two enzymes are highlighted in red and conservative mutations are indicated by red font. Arrows indicate break and splice points chosen for the chimera. (B) Structural alignment of KDM1A and KDM1B. KDM1A is shown in green and KDM1B is shown in blue. Inset shows ~90° rotation and close up of the aligned structures with TαA and TαB of the tower domain denoted and the KDM1B loop shown in orange. (C) Structural model of chKDM1AΔTower. AOD is shown in magenta, SWIRM domain is shown in green, and KDM1B loop is shown in orange.