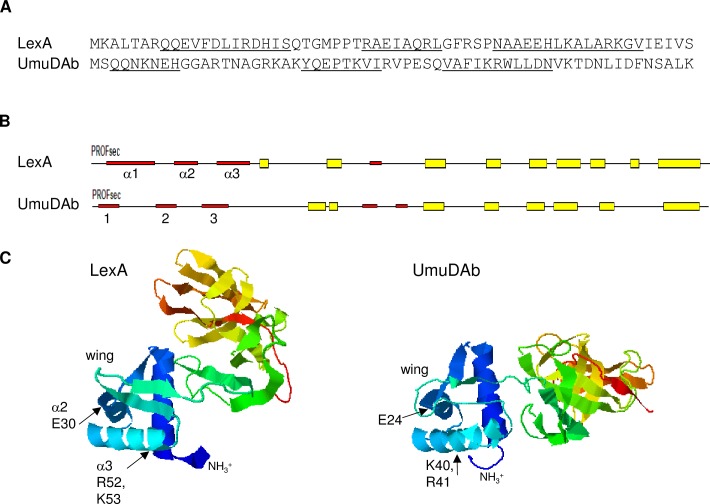
Fig 3. Modeling of the N-terminal domains of LexA and UmuDAb monomers.
(A) The N-terminal 60 amino acids of E. coli LexA and A. baylyi UmuDAb, showing underlined alpha-helical regions predicted by the Predict Protein server [28]. In LexA, helices α1–3 span amino acids 8–20, 28–35, and 41–55 [25]. For UmuDAb, helices are predicted to form from amino acids 3–9, 22–29, and 36–46. (B) Predicted secondary structures of LexA and UmuDAb, showing alpha-helices 1–3 represented by red thin bars and beta sheets represented by thick yellow boxes; predicted by the Predict Protein server. (C) I-TASSER modeling of LexA and UmuDAb, oriented to align the NTDs (in blue shading) and showing the wing of the wHTH structures. The interdomain linker between the LexA NTD and CTD is extremely flexible [4] and is likely responsible for the variation between the two proteins’ total orientations. Arrows point to some of the amino acids in the LexA α2 helix and α3 recognition helix that are required for DNA binding [4], and the similarly located sites of directed mutations in UmuDAb.