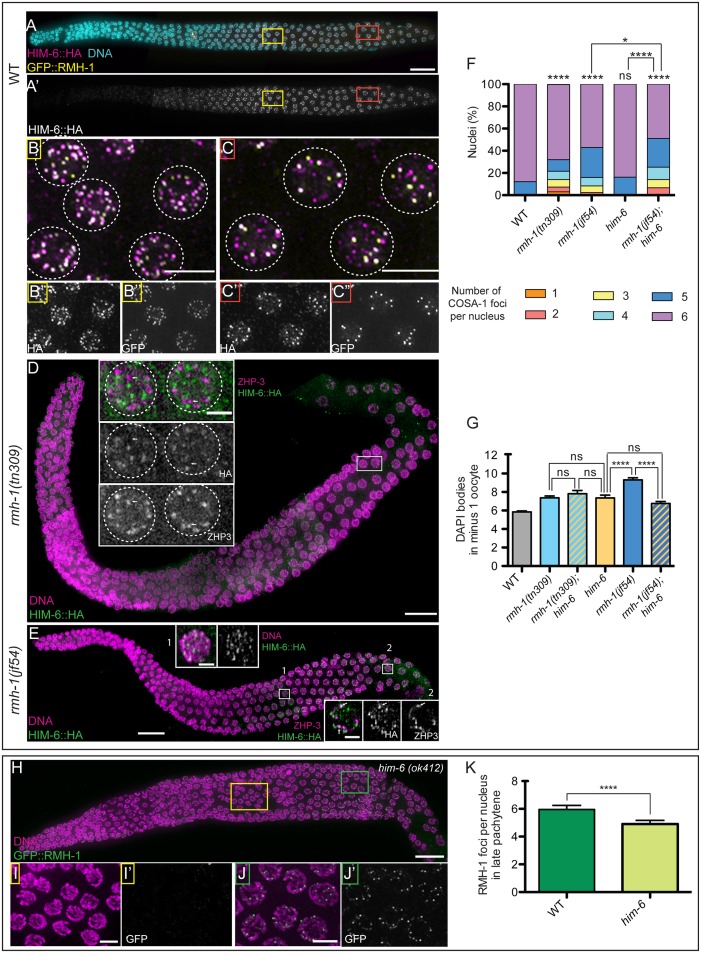
Fig 5. Separable and interdependent functions of RMH-1 and HIM-6 (BLM).
(A-C) Comparison of RMH-1::GFP and HIM-6(BLM)::HA localization in wild-type germ cells. (B–B′′) In mid pachytene (MP) nuclei, a high number of GFP::RMH-1 and HIM-6::HA foci are present, and most colocalize. (C–C′′) In late pachytene (LP), HIM-6 and RMH-1 colocalize in bright foci, but additional HIM-6 signals are present at other sites. (D) In rmh-1(tn309), HIM-6 foci are very small and faint (two gonads) or completely undetectable (three gonads). Inset: residual HIM-6 foci do not colocalize (white arrows) with ZHP-3. (E) In rmh-1(jf54), HIM-6 is still present in foci. Insets for MP (1) and LP (2); in (2), HIM-6 foci can be found in proximity of ZHP-3 at some putative CO-designated sites; however, most do not colocalize. (F) Stacked bar graph showing the percentage of nuclei containing a defined number of COSA-1 foci either in WT (n = 428), rmh-1(jf54) (n = 291), rmh-1(tn309) (n = 329), him-6(ok412) (n = 204), or rmh-1(jf54); him-6 (n = 177). The data presented for WT, rmh-1(jf54), and rmh-1(tn309) are the same as in Fig 4F. Difference in COSA-1 foci distribution was assessed by a Mann Whitney test: ns stands for not significant and **** p < 0.0001. (G) Quantification of the number of DAPI positive structures in the -1 oocyte for WT; single mutants (rmh-1(tn309)- him-6(ok412)- rmh-1(jf54)) and the double rmh-1(tn309); him-6 and rmh-1(jf54); him-6 (n = 15–36 nuclei per genotype). Data are represented as mean +/- SEM with ns (not significant) and **** p < 0.001. (H) In him-6(ok412), RMH-1 is not detectable in MP (I,I′), but bright RMH-1 foci are present in LP (J,J′), albeit quantification (K) indicates that their numbers are reduced relative to WT (p < 0.001). Data are represented as mean +/- SD. Scale bar 20 μm for gonads, 5 μm for pachytene nuclei insets, and 2.5 μm for single nuclei.