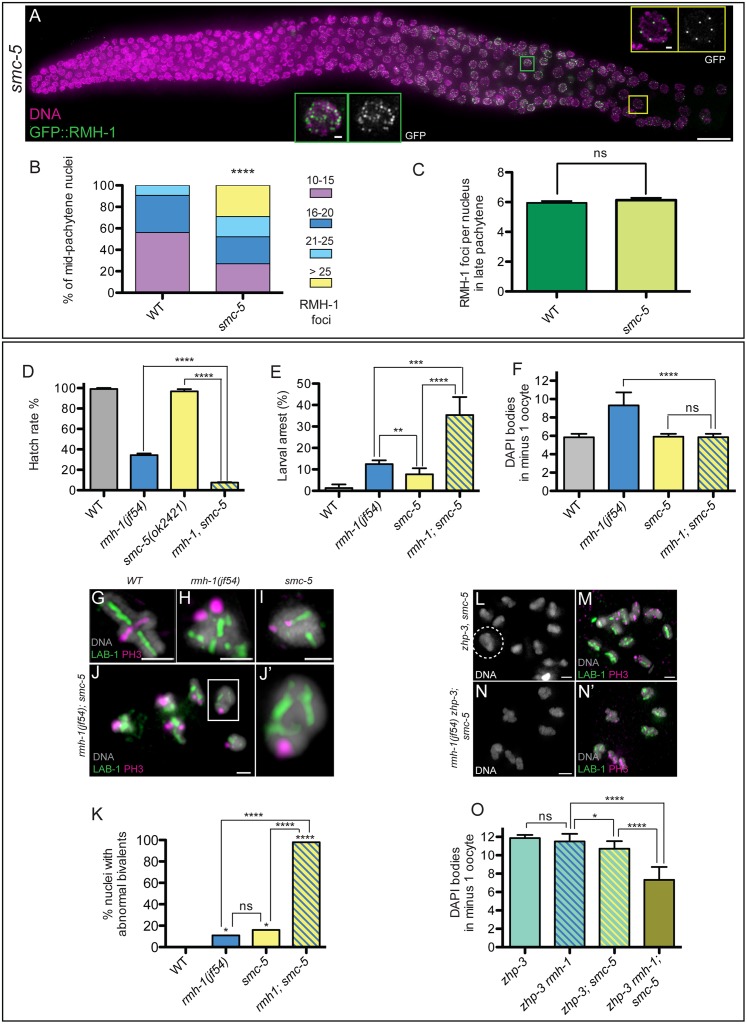
Fig 7. RMH-1 and SMC-5 cooperate to prevent accumulation of aberrant interhomolog connections.
(A) In smc-5(ok2421), RMH-1 foci increase in mid pachytene nuclei (MP) (green square), while in late pachytene (LP) (yellow square), a zone with fewer foci is still present, as in WT. (B) Quantification of RMH-1 foci in MP nuclei in WT (n = 205) and smc-5(ok2421) (n = 203). In the mutant, we frequently observe nuclei with more than 25 foci, never seen in the WT. Distribution of mid pachytene GFP::RMH-1 foci is significantly different between WT and smc-5 mutant (Mann Whitney test, **** p < 0.0001). (C) Quantification of RMH-1 foci in LP nuclei; data are represented as mean +/- SD with ns (not significant). Quantification of hatch rate (D), larval arrest (E), and DAPI bodies in diakinesis oocytes (F) for WT, rmh-1(jf54), smc-5(ok2421), and rmh-1(jf54); smc-5 (n = 35–45 hermaphrodites per genotype). Data are represented as mean +/- SD with ns (not significant) and * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. (G–J) Images of individual diakinesis bivalents stained for long arm and short arm markers. Both rmh-1(jf54) and smc-5 single mutants exhibit abnormally structured bivalents at low frequency (H,I). In rmh-1; smc-5, all diakinesis nuclei contain bivalents with abnormal structures; typical of these abnormalities is a side-by-side organization of the long arms of the bivalents (J′), presumably reflecting the presence of persistent interhomolog associations at NCO sites. (K) Quantification of the frequencies of diakinesis nuclei (-2 and -1) containing at least one abnormal bivalent (n = 13–25 nuclei per genotype). Data are represented as percentage with ns (not significant) and ** p < 0.01, *** p < 0.001, and **** p < 0.0001 (Chi2 test) (L–N) Images of chromosomes in diakinesis nuclei from zhp-3; smc-5 and rmh-1(jf54) zhp-3; smc-5 worms. Despite the absence of the canonical meiotic CO machinery component ZHP-3, fewer than 12 DAPI structures are observed in some zhp-3; smc-5–1 oocytes, indicating the presence of ectopic connections (L,M). Such ectopic connections occur at high frequency in the rmh-1(jf54) zhp-3; smc-5 triple mutant (N–N′). The quantification is presented in (O) with n = 13–36 oocytes per genotype. Data are represented as mean +/- SD with ns (not significant), * p < 0.05 and **** p < 0.0001.