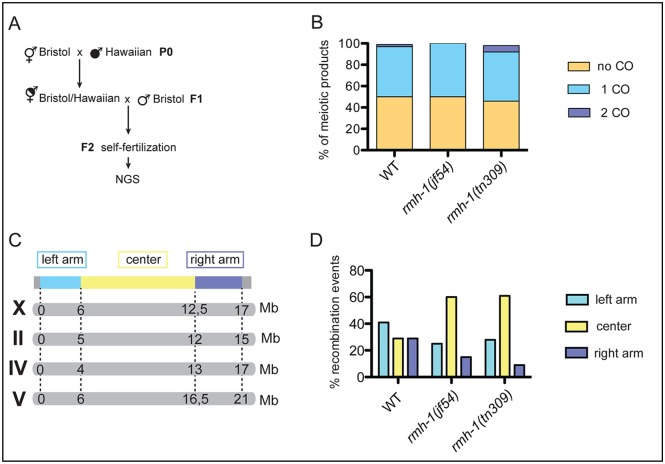
Fig 8. RMH-1 promotes the bias for CO formation on chromosome arms.
(A) Schematics of crosses to obtain the progeny of singled F2 individuals subjected to Next Generation Sequencing (NGS) for SNP analysis. White insert indicates the WT (Bristol) background, and black insert indicates the Hawaiian background. (B) Quantification of the overall recombination frequencies for assayed chromosomes; stacked bar graph indicates the fraction of meiotic products with zero, one, or two COs. For WT (n = 36 chromatids), for rmh-1(jf54) (n = 40 chromatids), and for rmh-1(tn309) (n = 45 chromatids). The frequency of COs was not found to be different between WT and both mutants (Chi2 test). (C) Scheme of the different chromosomes used during the recombination assay. The chromosome domains (left arm in blue, center in yellow, and right arm in purple) are correlated with the physical map of each chromosome. (D) Locations of the recombination events (assayed for chromosomes X, IV, and V) in WT (n = 17 COs: three events on X, four on II, four on IV, and six on V), for rmh-1(jf54) (n = 20 COs: 11 events on II and 9 on V), and rmh-1(tn309) (n = 21 COs: nine events on X, nine on IV, and three on V); also see the Experimental Procedures section. The relative distribution of COs in the center versus arm domains differed from the WT for tn309 (p = 0.046, Chi2 test) and for jf54, (p = 0.062, Chi2 test).