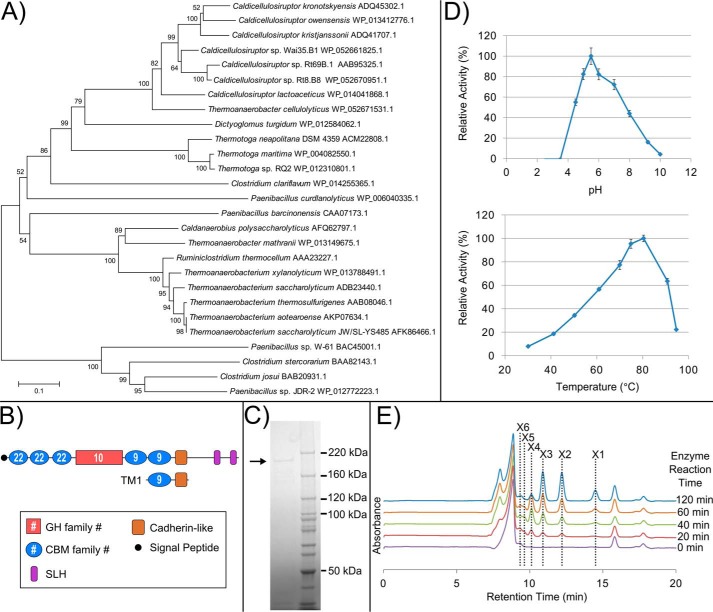
FIGURE 4.
Biochemical characterization of Calkro_0402. A, MEGA version 6 (95) was used to align, using MUSCLE (96), full protein amino acid sequences and construct an unrooted neighbor-joining phylogenetic tree of close blastp homologs to Calkro_0402 and previously characterized CBM22/GH10/CBM9-containing enzymes. Species and protein accession number are shown for each enzyme branch. B, domain organization for Calkro_0402 and TM1, which was used as the antigen for antibody production. Cadherin-like (pfam12733). C, purified full-length Calkro_0402 (∼180 kDa) produced in E. coli and purified by immobilized metal affinity and size exclusion chromatography. D, optimum pH at 70 °C and the optimum temperature at this optimum pH were determined for recombinant Calkro_0402 (0.02 mg/ml incubated 30 min). Error bars, S.D. (n = 3). E, oligosaccharides released from birchwood xylan by Calkro_0402. HPLC chromatograms for each enzyme reaction time are aligned with respect to retention time and graphed together. The location of the peaks for the standards xylose (X1), xylobiose (X2), xylotriose (X3), xylotetraose (X4), xylopentaose (X5), and xylohexaose (X6) are shown.