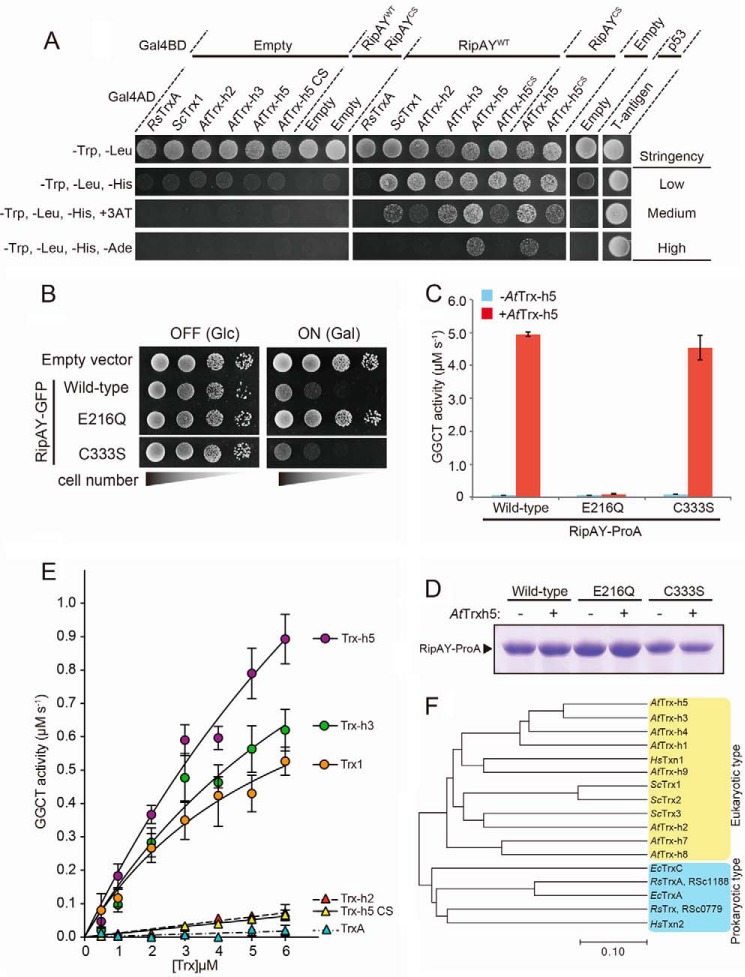
FIGURE 8.
Plant thioredoxins can bind to RipAY and activate GGCT activity of RipAY in an isoform-specific manner. A, yeast two-hybrid assay for interaction between RipAY and various thioredoxins on the plate with different stringency conditions. RipAY WT (RipAYWT) and a mutant form of RipAY (RipAYCS), in which the sole cysteine residue at position 333 was substituted by serine, were used as bait. R. solanacearum TrxA (RsTrxA), S. cerevisiae Trx1 (ScTrx1), A. thaliana Trx-h2, -h3, and -h5 (AtTrx-h2, AtTrx-h3, and AtTrx-h5), and a mutant form of Trx-h5 (AtTrx-h5CS), in which active site cysteine residues at position 39 and 42 were substituted by serine, were used as prey. B, yeast cells carrying empty vector or GAL1 expression vector of WT, active site mutant E216Q, or C333S mutant of RipAY were spotted on SD (−Ura) and SGal (−Ura) plates and cultured for 2 days and 3 days, respectively. C, GGCT activity of the beads bound with Protein A-tagged RipAYWT, RipAYE216Q, or RipAYC333S proteins expressed in E. coli incubated with 1.6 μm AtTrx-h5-His6 protein was measured by the Dug1-coupled method. Values represent the mean ± S.E. (error bars) (n = 3). D, Protein A-tagged RipAYWT, RipAYE216Q, and RipAYC333S proteins extracted from the beads shown in C were resolved on SDS-PAGE and visualized by Coomassie Brilliant Blue staining. E, effect of thioredoxin isoforms on activation of GGCT activity of RipAY. The GGCT activity of RipAY incubated with various concentrations of thioredoxin isoforms was measured by the Dug1-coupled method. Values represent the mean ± S.E. (n ≥ 3). F, phylogenetic analysis of thioredoxin isoforms from various organisms. The organisms, locus tag/gene name, and GenbankTM accession numbers (in parentheses) are as follows: R. solanacearum, RSc1188/RsTrxA (AL646052.1) and RSc0779/RsTrx (AL646052.1); E. coli, EcTrxA (M26133.1) and EcTrxC (U8594.1); H. sapiens, HsTxn1 (JQ313905.1) and HsTxn2 (DQ891579.2); S. cerevisiae, YLR043C/ScTrx1 (NM_001181930.1), YGR209C/ScTrx2 (NM_001181338.3), and YCR083C/ScTrx3 (NM_001178789.1); A. thaliana, At3g51030/AtTrx-h1 (NM_114963.4), At5g39950/AtTrx-h2 (AY113052.1), At5g42980/AtTrx-h3 (AY065098.1), At1g19730/AtTrx-h4 (BT004710.1), At1g45145/AtTrx-h5 (AY040028.1), At1g59730/AtTrx-h7 (BT0031540.1), At1g69880/AtTrx-h8 (BT003670.1), and At3g08710/AtTrx-h9 (BT0011728.1).