FIGURE 4.
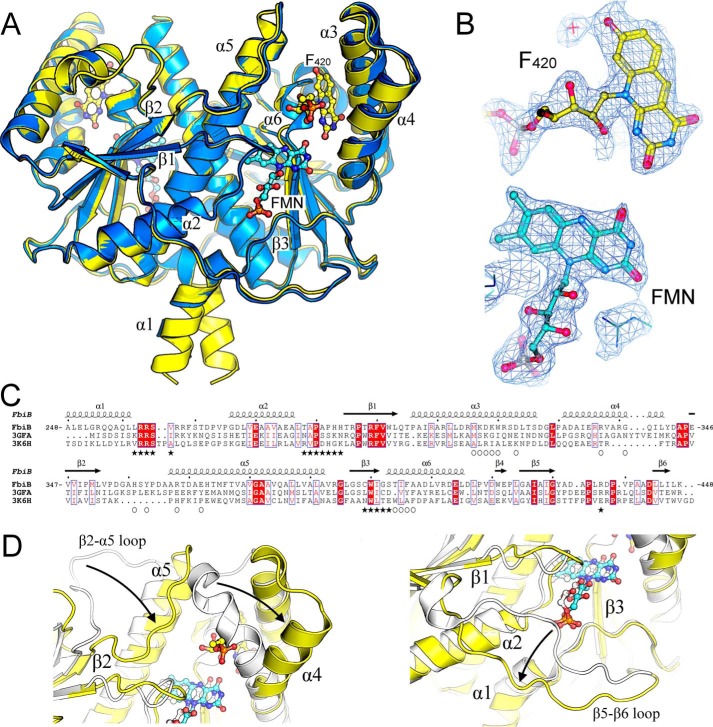
Crystal structure of the dimeric C-terminal domain of FbiB. A, overlay of F420-bound (yellow) and FMN-bound (blue) structures. F420 and FMN molecules are shown as ball-and-stick models with carbon atoms colored in yellow and cyan, respectively. Selected secondary structure elements are labeled. B, electron density (2Fo − Fo, omit map) surrounding F420 and FMN molecules in their respective structures. C, multiple sequence alignment of FbiB (C-terminal domain), the putative FMN-dependent nitroreductase from C. difficile (Protein Data Bank entry 3GFA), and a nitroreductase from A. tumefaciens (Protein Data Bank entry 3K6H). Secondary structure elements are shown for the FbiB structure, and residues involved in F420 and FMN binding are indicated by open circles and solid stars, respectively. Many residues involved in FMN binding appear conserved across the three structures, whereas those involved in F420 binding are not conserved. D, overlay of enlarged view of FbiB (yellow) and Protein Data Bank entry 3GFA (white). Incorporation of an F420 binding site (left) produces a large shift in the top of helix α4 and in the β2-α5 loop compared with the C. difficile homolog structure. Another large difference occurs at the bottom of the molecule (right), involving the β5-β6 loop, that opens up in FbiB to produce the binding site for the phosphate group of FMN.