FIGURE 4.
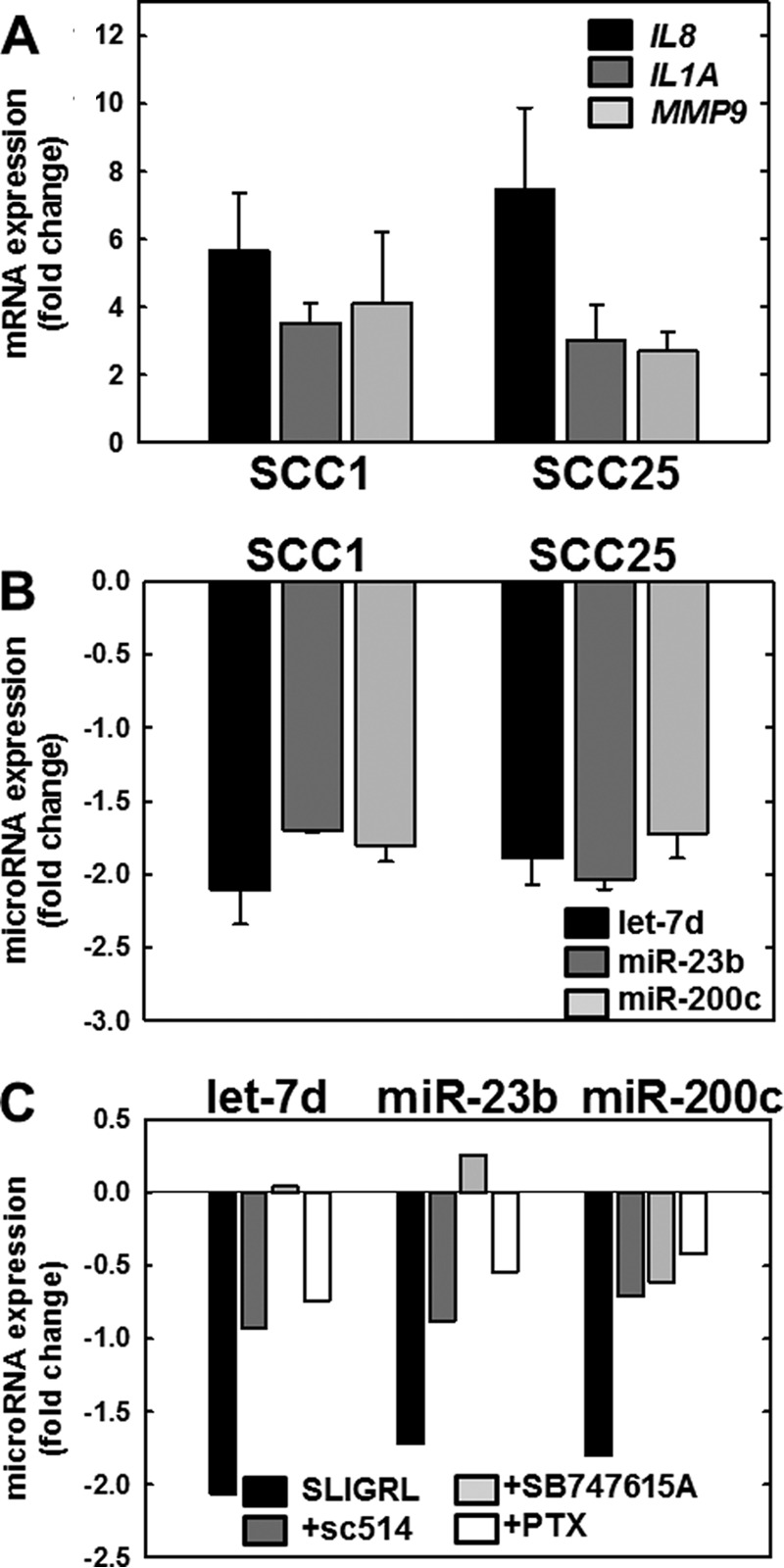
PAR-2 activation regulates expression of mRNAs and microRNAs in OSCC cell lines. A, PAR-2 activation increases pro-inflammatory mRNAs. PAR-2 was activated in SCC1 or SCC25 cell lines, as indicated, by incubation with SLIGRL-NH2 (20 μm, 2 h) followed by immediate RNA isolation and processing for RT-qPCR as described under “Materials and Methods.” Data shown are fold-change in expression of IL8 (black bar), IL1A (dark gray bar), and MMP9 (light gray bar) relative to untreated cells. Experiments were repeated in triplicate; error bars show S.E. B, PAR-2 activation decreases anti-inflammatory microRNAs. PAR-2 was activated in SCC1 or SCC25 cell lines, as indicated, by incubation with SLIGRL-NH2 (20 μm, 2 h) followed by immediate RNA isolation and processing for RT-qPCR as described under “Materials and Methods.” Data shown are fold-change in expression of let-7d (black bar), miR-23b (dark gray bar), and miR-200c (light gray bar) relative to untreated cells. Experiments were repeated in triplicate; error bars show S.E. C, PAR-2-mediated microRNA suppression is reversed by Nf-κB inhibitors. Cells were incubated with vehicle (control, black bar) or pretreated overnight with inhibitors as indicated: sc-514 (100 μm, dark gray bar), SB747651A (20 μm, light gray bar), or pertussis toxin (PTX) (1 μg/ml, 8.5 nm white bar) prior to activation of PAR-2 with SLIGRL-NH2 (20 μm, 2 h) in the continued presence of inhibitor. RNA was isolated immediately and used as template for RT-qPCR. Data shown are fold-change in expression of let-7d, miR-23b, and miR-200c relative to SLIGRL-NH2-activated cells without the addition of an inhibitor. Experiments were performed in duplicate.
