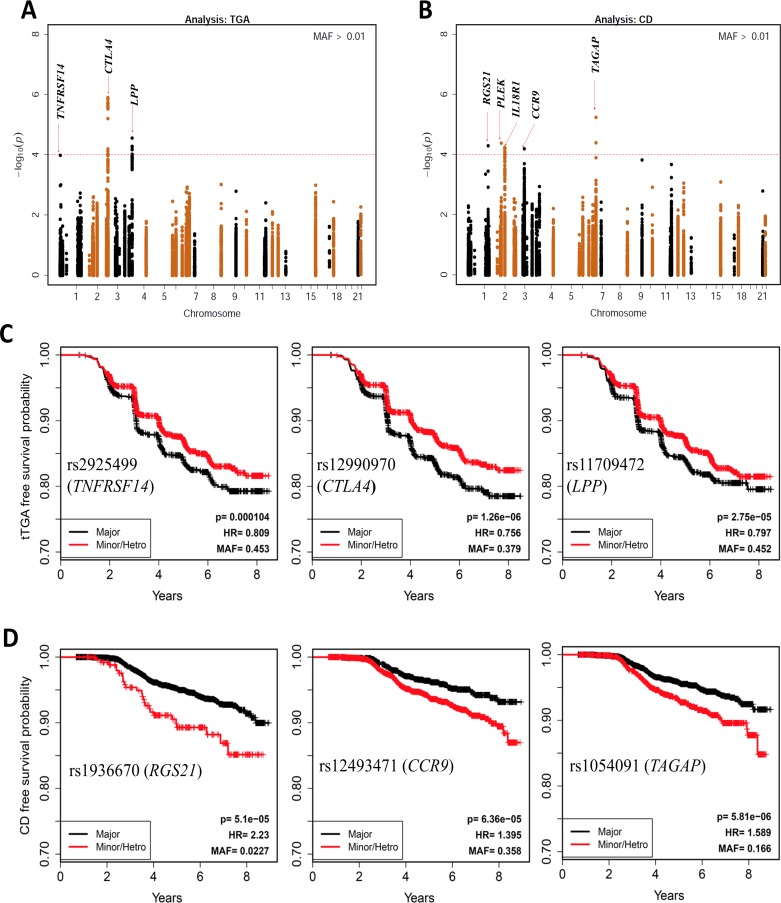
Fig 1. SNPs in the previously reported celiac disease associated regions.
Manhattan plot of P-values on the −log10 scale for SNPs (±400kb) previously associated with celiac disease (A) and persistent tissue transglutaminase autoantibody (tTGA) positivity (B). HRs and p-values are calculated using three possible genotypes and adjusted for family history of celiac disease, HLA-DR-DQ genotype, gender, HLA-DPB1, population stratification (ancestral heterogeneity) and country of residence (as strata). The red dashed line represents p = 1x10−4. Kaplan-Meier plots of the three most significant SNPs associated with celiac disease (C) and tTGA (D) are plotted by dividing the subjects in two groups: (i) Major homozygous (black curves) and (ii) Heterozygous combined with minor homozygous (red curves).