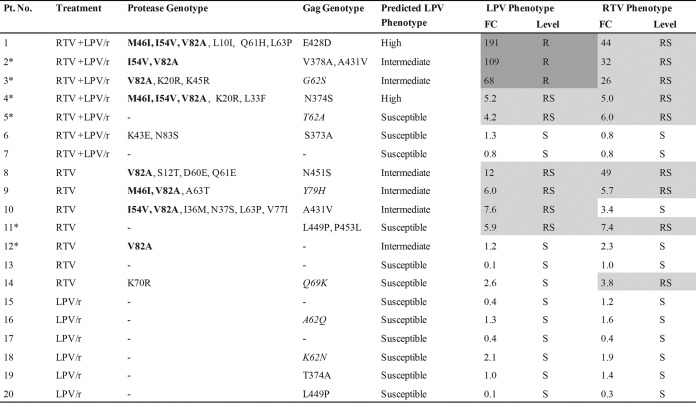
FIG 1.
Genotypic and phenotypic resistance of pediatric patient samples to LPV and RTV at treatment failure. The treatment histories of the patients are shown, where RTV + LPV/r indicates the receipt of both RTV as a single PI and LPV/r at some point, RTV indicates the receipt of RTV as a single PI only, and LPV/r refers to the receipt of LPV/r only. The Stanford HIV drug resistance database was used to predict the phenotype. Asterisks, the six patients for whom discordance between the predicted and the actual phenotype was found. Levels of drug resistance were classified as follows: sensitive (S; no shading), reduced susceptibility (RS; light gray shading), and resistant (R; dark gray shading). The assay cutoffs used are as follows: for sensitive, a <3.6-FC in the EC50 for LPV and a <3.8-FC in the EC50 for RTV; for reduced susceptibility, a 3.6- to 55-FC in the EC50 for LPV and a >3.8-FC in the EC50 for RTV; and for resistant, a >55-FC in the EC50 for LPV. Protease amino acid changes in bold represent major PI mutations. Gag amino acid changes in italics represent those at non-CSs that were previously shown to be associated with PI exposure (28).