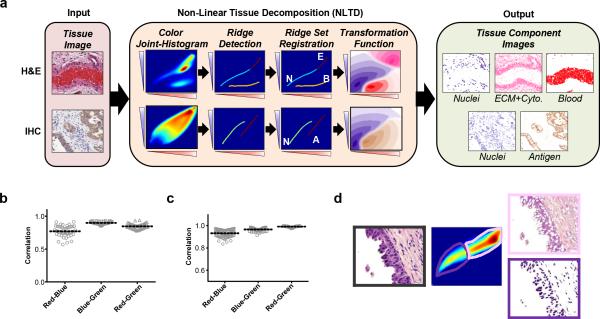
Figure 1. Brief overview of NLTD approach.
(a) NLTD applied to an image of a hematoxylin and eosin (H&E) stained section (top) and immunohistochemically (IHC) stained image (bottom). Shown are a typical H&E image of a small artery, exhibiting multiple tissue components (nuclei (N), ECM-rich and cytoplasm (E), blood (B)) and a typical IHC image, stained for LINE-1 ORF1p expression31, exhibiting two tissue components (antigen (A) and nuclei (N)). The NLTD method is schematically shown in center. Briefly, the red-blue joint-histogram is first segmented to identify each region in the red-blue color space. The x-axis corresponds to each red color, the y-axis shows each blue-color, and the color-axis represents the frequency of each discrete color combination. Ridges for each tissue component are overlaid, on the RBJH. The ridge set is registered and transformed to yield the pseudo-colored transformation function for each component. The pseudo-colored grayscale images are shown for the nuclei, non-nuclei, and blood components (purple, pink, and red, respectively) in the far right box. (b-c) Grayscale correlation values for the red-blue joint histogram, blue-green joint histogram, and red-green joint-histogram, with a value of 1 corresponding to a completely correlated colorset. (b) Pancreatic cancer H&E dataset (n=45). (c) Ovarian immunohistochemistry dataset (n=81). (d) Separation of red-blue color space into individual tissue components: nuclei (purple box), extracellular matrix and cytoplasm rich (pink box).