Figure 7. Analysis of SRF Target Genes.
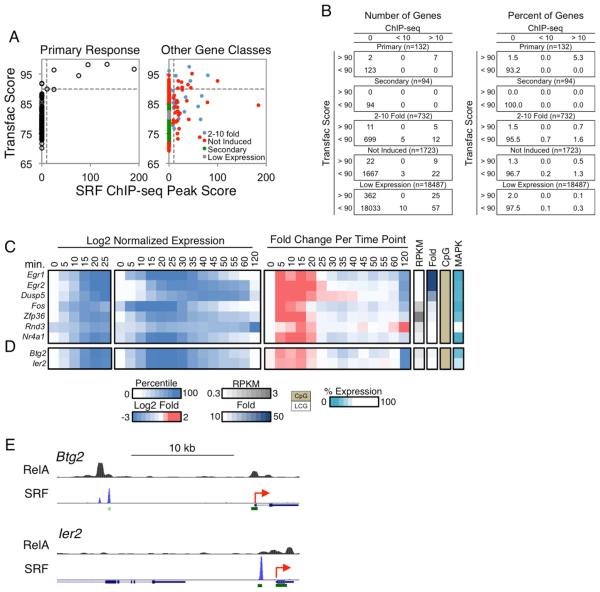
(A) Scatterplots comparing the Transfac PWM scores of SRF binding motifs (y-axis) versus the SRF ChIP-seq peak scores (x-axis) in the promoters (−500 to +150) of the PRGs (left) and all remaining genes in the genome (right) is shown. The genes in the latter graph were divided into categories as in Figure 4A. The horizontal and vertical dashed lines indicate the SRF motif (90) and ChIP-seq peak (10) thresholds.
(B) Tables are shown indicating the distribution of genes from panel (A), for both numbers (left) and percentages (right) of genes.
(C) Log2 normalized expression values from 0-25 min (first panel), 0-120 min (second panel), and the fold induction relative to the expression level at the previous time point (third panel) are shown for the seven putative SRF target genes. To the right are columns indicating the basal expression level, fold induction magnitude, promoter CpG content, and MAPK dependence for each gene.
(D) Two genes that exhibited similar activation kinetics as the putative SRF target genes are shown, with the same layout as in Figure 7C.
(E) The two genes from panel (D) were examined on UCSC Genome Browser to identify distal SRF binding peaks. RelA binding peaks were also examined for these genes. The TSSs of the genes are indicated as red arrows, and the green rectangles indicate CpG islands. See also Figures S6 and S7.