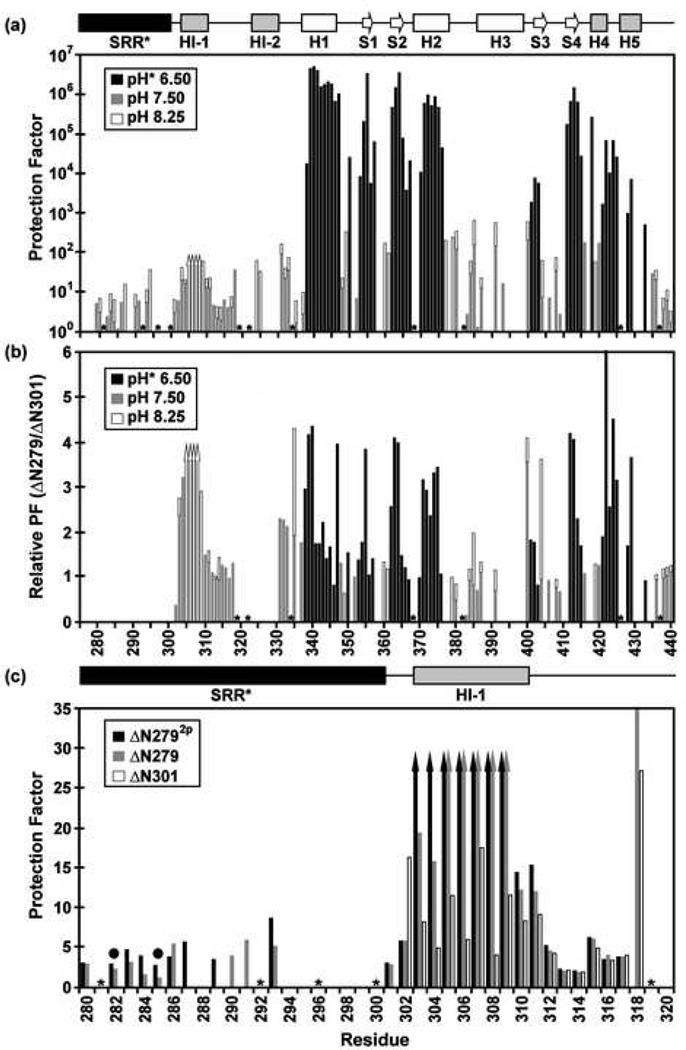
Fig. 4.
Amide HX measurements provide insights into the structure, stability, and dynamics of the Ets-1 deletion fragments (500 mM NaCl, 28 °C). (a) The comprehensive backbone amide HX protection factors of ΔN279, determined from slow H2O-D2O exchange (pH* 6.50, black) and fast H2O-H2O CLEANEX (pH 7.50, gray; pH 8.25, white) measurements. Residues 305–308 exchanged too slowly to be characterized by CLEANEX, yet too fast to be detected after transfer into D2O buffer, and thus their protection factors fall within the range of ~ 30 to 200 (up arrows). The SRR* is predominantly unstructured, showing little protection against HX. The inhibitory helices HI-1 and H1-2, as well as the DNA recognition helix H3, undergo relatively facile HX. (b) However, the presence of the SRR* stabilizes the inhibitory module and ETS domain against HX by up to ~ 4 fold. Shown are relative protection factors for ΔN279 versus ΔN301, obtained from H2O-D2O exchange (pH* 6.5, black) and/or CLEANEX measurements (pH 7.5, gray; pH 8.25, white). (c) Phosphorylation of S282 and S285 further stabilizes the N-terminal inhibitory helix of ΔN279 against amide HX. Shown are the amide protection factors of residues 280–320 in ΔN2792p (black), ΔN279 (gray), and ΔN301 (white), acquired from CLEANEX exchange experiments at pH 7.5. Up arrows indicate minimal estimated values. Dark circles indicate the phosphoacceptor serines, and missing data correspond to prolines (*) or amides with weak or overlapping NMR signals. Slow amide HX kinetics, which require measurements over many months, were not carried out with ΔN2792p.