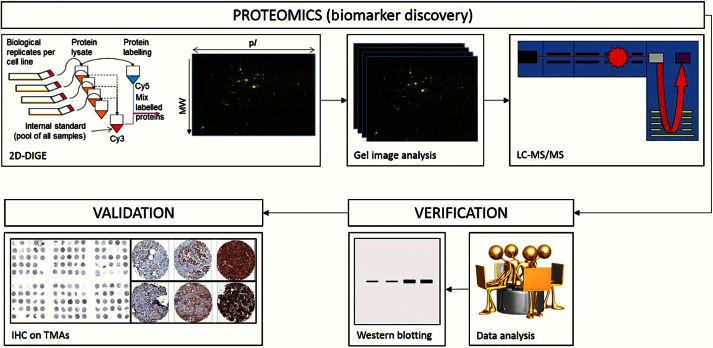
Figure 1.
Simplified flow diagram showing key proteomic steps used for discovery of novel biomarkers for risk of bone metastasis development. Proteins extracted from multiple independent cultures of cell lines were labeled using Cy5 fluorescent dye while an internal standard was labeled with Cy3. Separation of proteins (by isoelectric point and molecular weight) and image capture (fluoresence densitometry) creates protein array images that can be compared so as to detect differential protein expression between cell line types and replicates (intensity of fluoresence). Proteins of higher expression in bone-homed cell lines were excised from silver-stained two-dimensional difference gel electrophoresis gels, reduced to peptides, and analyzed using tandem mass spectrometry. Identified proteins were assessed for known/reported relevance to breast cancer and/or bone metastasis prior to selection for validation of expression on breast cancer tissue microarrays. 2D-DIGE = two-dimensional difference gel electrophoresis; IHC = immunohistochemistry; LC/MS/MS = liquid chromatography/mass spectrometry/mass spectrometry; TMA = tissue microarray.