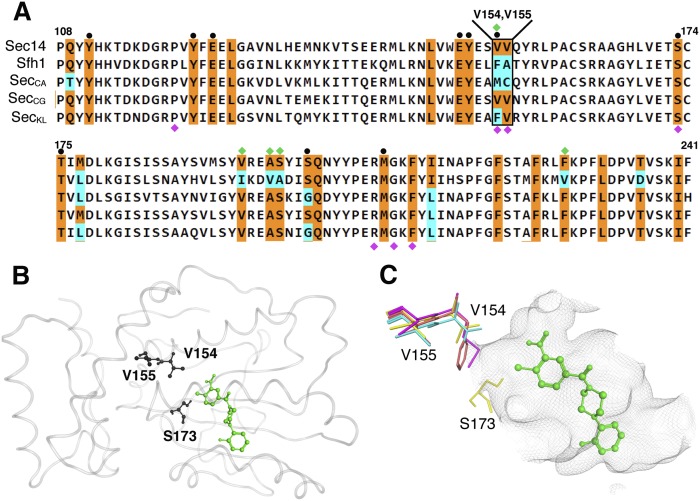
Fig. 4.
The Sec14 VV motif is not conserved among closely related Sec14 PtdIns/PtdCho-transfer proteins. A: Shown is a partial primary sequence alignment of S. cerevisiae Sec14 with Sfh1 and Sec14 PITPs from C. albicans, C. glabrata, and K. lactis. The residues corresponding to the lipid-binding cavity are highlighted in color. Residues conserved between S. cerevisiae Sec14 and other Sec14-like proteins are colored in orange with nonconserved residues in cyan. At top, the Sec14 VV motif is identified. For reference, PtdCho-binding barcode residues (black circles) and residues altered to produce the Sfh16X mutant (green diamonds) are also highlighted at top. Labeled at bottom are residues where substitutions confer NPPM resistance to Sec14 (magenta diamonds). B: The S. cerevisiae Sec14::NPPM 6748-481 dock model is shown. Protein is rendered in gray transparent loop with residues V154, V155, and S173 shown in black ball and stick render. NPPM 6748-481 is highlighted in green ball and stick mode. C: A magnified view shows the spatial orientation of the Sec14 V154V155 side chains (in stick render) relative to the model NPPM 6748-481 pose (in green ball and stick mode), residue S173 is indicated, as is the Sec14 hydrophobic pocket surface (rendered as a gray wire-mesh surface). The cognate VV motif residues from S. cerevisiae (yellow), C. albicans (magenta), C. glabrata (cyan), and K. lactis (orange) are overlayed.