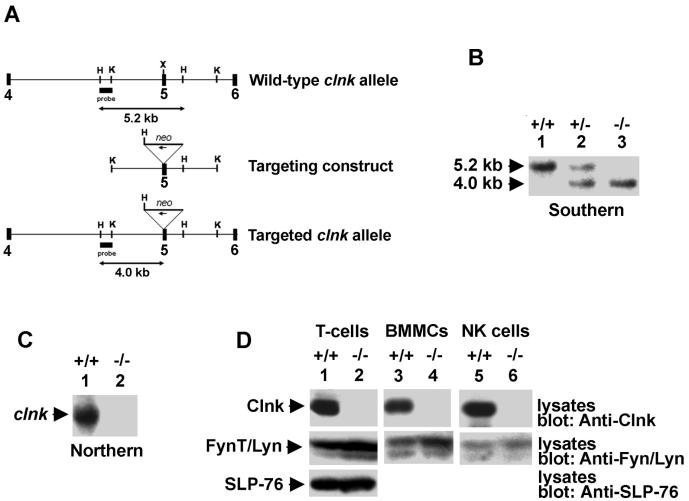
FIG. 1.
Molecular and biochemical characterization of Clnk-deficient mice. (A) clnk gene organization and targeting construct. A partial structure of the mouse clnk gene (exons 4 to 6) and a depiction of the targeting construct are shown. The position of the DNA fragment used as a probe for Southern blotting is shown. This probe hybridizes to a 5.2-kb HindIII fragment in clnk+/+ mice and to a 4.2-kb HindIII fragment in clnk−/− animals. (B) Southern blot depicting wild-type, heterozygous, and homozygous targeted alleles. Genomic DNA from mouse tails was digested with HindIII and tested by Southern blotting by using the probe depicted in panel A. (C) Northern blot. RNA was isolated from BMMCs derived from clnk+/+ and clnk−/− mice and probed by Northern blotting by using a full-length clnk cDNA as a probe. (D) Immunoblots. Lysates were prepared from anti-CD3 MAb-stimulated T cells (day 5 of IL-2 cultures), BMMCs, and NK cells. The presence of Clnk and that of SLP-76 were detected by immunoblotting by using polyclonal rabbit anti-Clnk (top panels) and anti-SLP-76 (bottom panel) sera, respectively. Equal loading was confirmed by reprobing the immunoblot membranes (middle panels) with anti-FynT (lanes 1 and 2) or anti-Lyn (lanes 3 to 6) antibodies.