Abstract
A phenol-degrading bacterium strain PA was successfully isolated from the effluent of petrochemical wastewater. Based on its morphological, physiological and biochemical characteristics, the strain PA was characterized as a Gram-negative, strictly aerobic, nonmotile and short rod-shaped bacterium that utilizes phenol as a sole carbon and energy source. 16S rDNA sequence analysis revealed that this strain is affiliated to Acinetobacter calcoaceticus in the group of Gammaproteobacteria. The strain was efficient in removing 91.6% of the initial 800 mg∙L−1 phenol within 48 h, and had a tolerance of phenol concentration as high as 1700 mg∙L−1. These results indicated that A. calcoaceticus possesses a promising potential in treating phenolic wastewater.
Keywords: phenol-degrading, biodegradation, Acinetobacter calcoaceticus
1. Introduction
The massive production and application of phenol in industrial activities make it a major environmental pollutant in most wastewater various facilities, such as oil refineries, coking plants, pharmaceuticals and plastic industries [1,2]. Many aquatic organisms, including microorganisms, plants and fishes, pose a risk of mutagenic, teratogenic and carcinogenic effects in the environments that contain phenol pollutants. Thus, phenol has been classified as a highly hazardous chemical [3], and has been included in the list of priority pollutants by the U.S. Environmental Protection Agency [4]. Finding an efficient method to remove phenol effectively has attracted increasing concern for the environmental remediation and the health welfare of human beings.
Physico-chemical methods, such as ultra violet, ozonation, hydrogen peroxide, Fenton’s reagent, or a combination of these methods, have been employed to eliminate phenol in industrial wastewater [5,6]. These methods were effective in removing phenol but were complex and costly. Hence, the development of improved technologies capable of degrading phenol in the environments is necessary. Alternatively, the biodegradation of phenols is an environmentally friendly and cost-effective technology currently preferred to reduce phenol compounds [7].
Based on the theory that a few microorganisms can utilize phenol as the sole source of carbon and energy [8,9], numerous bacterial species have been isolated and characterized as phenol-degrading microorganisms [10,11]. These bacterial species include Pseudomonas putida [12], Rhodococcus erythropolis [13], Bacillus subtilis [14], Bacillus brevis [11], Serratia marcescens [15], Arthrobacter citreus [16], Alcaligenes faecalis [17], Sphingomonas [18], and Acinetobacter [19,20,21]. Native microbial species have been reported to be more adaptive and capable of out-competing non-indigenous microorganisms in the remediation of special polluted environments [22]. Therefore, identifying new phenol-degrading bacteria is necessary for the bioremediation of the phenol-contaminated environments in various regions.
In the current study, a bacterial strain capable of degrading phenol was isolated from the petrochemical wastewater in South China. The strain PA was affiliated to Acinetobacter calcoaceticus in the group of Gammaproteobacteria. The degradation efficiencies of phenol for the strain were then examined. In addition, the growth limitation of the strain in elevated phenol concentrations was evaluated.
2. Materials and Methods
2.1. Chemicals, Media and Phenolic Wastewater Sample
All chemicals used were analytical reagents. The minimal salt media (MSM) and Luria-Bertani (LB) media were used in present study. The MSM contained KH2PO4 0.5 g, K2HPO4 0.5 g, CaCl2 0.1 g, NaCl 0.2 g, MgSO4·7H2O 0.5 g, MnSO4·7H2O 0.01 g, FeSO4·7H2O 0.01 g, NH4NO3 1.0 g per liter. The LB media was composed of tryptone 10 g, yeast extract 5 g and NaCl 5 g per liter. Deionized, distilled water was used for the experiments. The phenolic wastewaster samples along with activated sludge were collected in a biological treatment system from a petrochemical company in Guangdong, China. The phenol concentration in the wastewater was approximately 100 mg·L−1.
2.2. Enrichment and Isolation of Phenol-Degrading Bacteria
The phenolic wastewater samples collected from an oil refinery effluent in South China was inoculated into flasks containing MSM for the enrichment culture. Phenol was supplemented in the media as the sole carbon source, and the various concentrations of phenol were 200, 500, 800, 1100, 1400, 1700, 2000 and 2500 mg·L−1. The enriched culture observed with more biomass (in the flask with 800 mg·L−1 phenol) was further transferred into a freshly prepared enrichment media with higher concentrations of phenol (increased from 800 mg·L−1 to 2500 mg·L−1). The final enriched media were diluted serially and spread on LB agar plates supplemented with phenol (500 mg·L−1). The plates were incubated at 30 °C and single colonies with morphological differences were selected and streaked on new plates. The resulting isolates were stored at 4 °C for further study.
The morphological properties of the isolated colonies were observed by optical microscopy. The typical physiological and biochemical characteristics of the phenol-degrading bacteria strains, such as Gram’s staining, motility, starch hydrolysis, and gelatinase [23] were systematically performed according to Bergey’s manual of determinative of bacteriology [24]. Indole test, methyl red test and hydrogen sulfide test were also analyzed, as described previously [25].
2.3. Electron Microscopy
SEM (scanning electron microscopy): Bacterial cell morphologies of phenol-degrading strains were examined by scanning electron microscope S-3000N (Hitachi, Tokyo, Japan). For the preparations, cells were fixed in 0.1 M phosphate buffer solution (PBS) containing 2.5% (v/v) glutaraldehyde at 4 °C for 5 h. The samples were then washed with PBS three times (10 min each time). For dehydration, the samples were treated with ethanol serials (30%, 50%, 70%, 90%, and 100%, v/v) for 20 min each. Dehydrated cells were filtered through a 0.2 μM polycarbonate filter, dried with a CO2-critical point dryer, coated with gold and observed subsequently by SEM at 20 kV.
TEM (transmission electron microscopy): The strain PA cells were harvested by centrifugation, washed with distilled water, and then placed onto carbon-coated nickel grids. After the cells were settled on the grid for 10 min, the excess liquid was drained off with filter paper, and the samples were air-dried. Subsequently, observations were preformed with a transmission electron microscope H-7650 (Hitachi, Tokyo, Japan) operating at 80 kV.
2.4. Characterization by 16S rDNA and Phylogenetic Analysis
The genomic DNA of strain PA was extracted as previously described [26]. DNA was used as template to amplify bacterial 16S rDNA with universal primers 27F (5′-AGAGATTGATCCTGGCTCTG-3′) and 1492R (5′-GGTTTCCTTGTTACGACAT-3′) on a Mastercycler gradient thermocycler (Eppendorf, Hamburg, Germany). The amplification reaction was performed in 25 μL volume PCR buffer containing 1.5 mM MgCl2, 0.2 mM of each deoxynucleoside triphosphate, 1 μM each of the forward and reverse primers. An initial denaturation step of 5 min at 94 °C was conducted, followed by 35 cycles of 94 °C for 30 s, 55 °C for 45 s and 72 °C for 90 s. The procedure was completed with a final elongation step at 72 °C for 10 min. The PCR products were sequenced, and the sequences were compared with bacterial 16S rDNA sequences in GenBank using the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) program [27]. Neighbor-joining phylogenetic trees [28] were constructed using the Molecular Evolutionary Genetics Analysis (MEGA) program version 4 [29]. The reliability of phylogenetic reconstructions was estimated through bootstrap analysis (1000 replicates). The 16S rDNA sequence of the strain PA was available under GenBank accession numbers KT878384.
2.5. Phenol Degradation
The culture of strain PA was prepared and adjusted to an optical density at 600 nm (OD600) of 1.0, then the final concentration of 2% (v/v) inoculums were inoculated into the flasks containing MSM media with phenol as sole carbon source. The range of phenol concentrations was increased from 200 to 1700 mg·L−1. The flasks were incubated at 30 °C with 150 rpm for 2 days. Samples were collected periodically to measure the biomass and the phenol degradation. The biomass contents were monitored spectrophotometrically by measuring absorbance at 600 nm. The phenol concentrations were determined by using 4-aminoantipyrine in the colorimetric assay, according to standard methods reported by the American Public Health Association [30].
3. Results and Discussion
3.1. Isolation and Characterization of Phenol-Degrading Strains
The wastewater and sludge samples collected from an oil refinery effluent in South China were inoculated in the medium containing phenol for the enrichment and isolation of phenol-degrading bacteria. After three weeks of enrichment and one week of strain isolation, a total of 10 isolates were obtained after 24 h growth on the LB agar plates with 100 μL of a 105–106 fold dilution of enrichment culture. All these stains utilized phenol as the sole carbon source and energy, and 1 of the 10 isolates exhibited more growth in phenol-containing media than the others. The outstanding isolate was named as phenol-degrading strain PA and was applied in the following study.
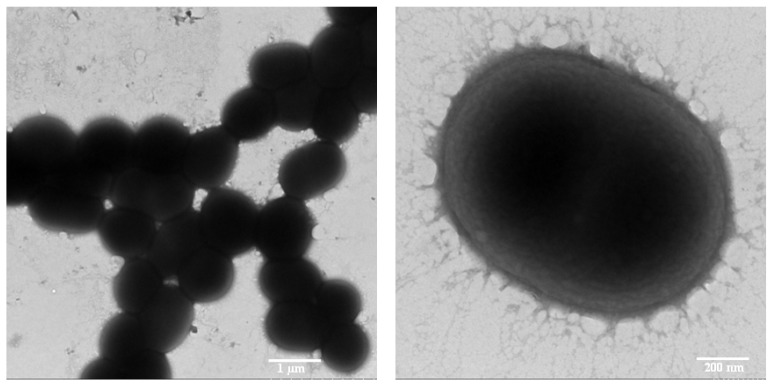
The strain PA was a Gram-negative and short rod-shaped bacterium with a cell size approximately 1.2 μM in length and 0.85 μM in diameter under the microscope (Figure 1). The biochemical characteristics of strain PA were determined, and biochemical tests showed that this strain was a catalase-positive, oxidase-negative bacterium (Table 1). This strain could grow at temperatures range of 20 °C–45 °C and a wide range of pH 5–11. The optimum growth was at the condition of 30 °C and pH 8.0.
Figure 1.
Transmission electron micrograph of Acinetobacter calcoaceticus PA.
Table 1.
Morphological and biochemical characteristics among Acinetobacter strains.
| Characteristics | Acinetobacter | ||
|---|---|---|---|
| Strain PA (This Paper) | PND-4 [31] | PND-5 [31] | |
| Color of colonies | white | milk white | milk white |
| Morphology | short rod | ND | ND |
| Motility | − | ND | ND |
| Gram straining | − | − | − |
| Aerobic growth | + | + | + |
| Starch hydrolysis | − | − | − |
| Catalase activity | + | + | + |
| Gelatin hydrolysis | − | − | − |
| Indole production | + | − | − |
| Methyl red | + | − | − |
| Hydrogen sulfide test | + | ND | ND |
ND: not determined.
3.2. Identification and Phylogenetic Analysis
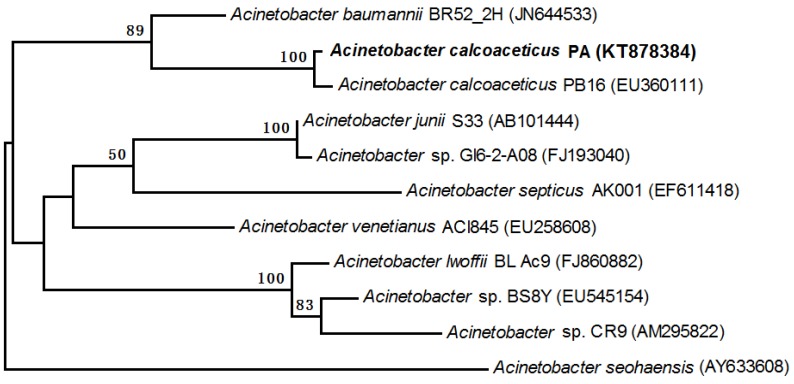
The 16S rRNA gene sequences of PA were sequenced and used to construct a phylogenetic tree for further analysis. The partial sequence of 16S rRNA gene was a continuous stretch of 1424 bp. The similarities between the PA sequence and the bacterial sequences deposited in the GenBank databases were calculated, and the PA sequence showed 99% similarity to that of A. calcoaceticus PB16. The phylogenetic analysis (1400 unambiguous bases aligned) revealed that the strain was classified in the Acinetobacter genera, which belong to the Gammaproteobacteria sub-phylum. Based on the neighbor-joining methods, a phylogenetic tree was constructed which indicated that the closest relative of strain PA was A. calcoaceticus PB16 (Figure 2). Therefore, the strain PA was identified and affiliated with A. calcoaceticus.
Figure 2.
Phylogenetic relationship based on the 16S rRNA gene sequences of strain PA and related organisms from the GenBank database. Bootstrap values were calculated from 1000 replications of Kimura 2-parameter, and bootstrap values higher than 50% were shown. The scale bar represents 0.002 changes per sequence position.
The Acinetobacter species were versatile in the biodegradation of various pollutants because of its highly hydrophobic cell surface which is adhesive to solid surfaces [32]. A. calcoaceticus is known to be a Gram-negative aerobic bacteria utilizing phenol as the sole source of carbon and energy [8,9]. A. calcoaceticus PHEA-2, isolated from phenol polluted wastewater in China [33], has been used for the complete genome analysis because of its high ability to effectively degrade phenol in the bioremediation of phenol-polluted wastewater [34].
3.3. Phenol Biodegradation by Acinetobacter Calcoaceticus PA
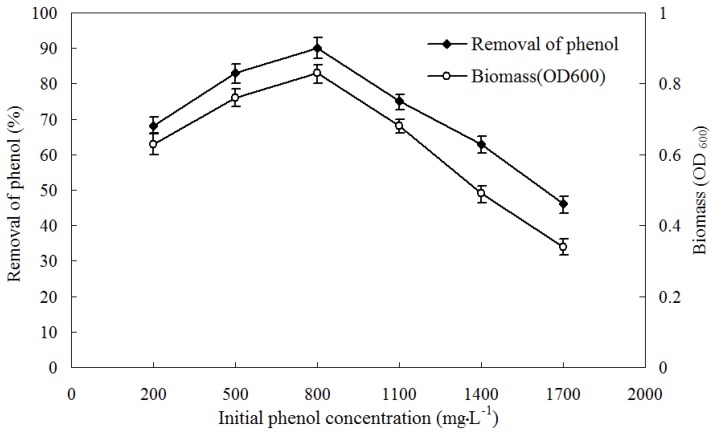
The phenol-degradation characteristics and biomass of A. calcoaceticus PA at various initial concentrations of phenol (200–2500 mg·L−1) were determined by monitoring phenol concentration and cell growth at OD600 periodically. The maximum biomass and degradation of phenol were observed at the initial phenol concentration of 800 mg·L−1 (Figure 3). An inhibitory effect showed that the biomass growth and the degradation of phenol were declined with the elevated initial phenol concentration higher than 800 mg·L−1. The removal rates of phenol were above 75% at the initial phenol concentration ranging from 500 to 1100 mg·L−1. While there was no growth of phenol-degrading bacteria when the initial phenol concentration was higher than 2000 mg·L−1. The strain could grow on phenol up to a concentration of 1700 mg·L−1 with the degradation rate of 46.2%.
Figure 3.
Profile of bacterial cell growth and phenol degradation at various initial concentrations of phenol.
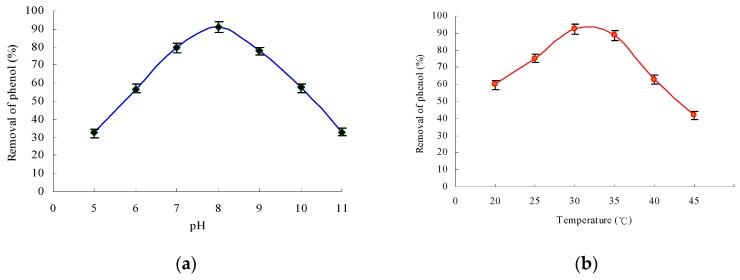
The effects of factors such pH values and temperature on the degradation were investigated. The bacterial strain could grow within a range of pH 5–11 (Figure 4a), and the degradation of phenol was above 78% in the range of pH 7–9. The optimum pH for phenol degradation was 8.0. These results showed that the bacterial growth of strain PA and the degradation of phenol (above 70%) were favored at temperatures of 25 °C–37.5 °C (Figure 4b). The biomass and phenol degradation reached the maximal values at a temperature of 30 °C, and both values showed only a slight change when the temperature was increased to 35 °C. On the contrary, the phenol degradation declined sharply when the temperature reached 40 °C and over. Therefore, the optimal temperature for the growth of strain PA was 30 °C. These growth conditions of A. calcoaceticus PA for phenol degradation were similar to those of other Acinetobacter reported previously [18,35,36].
Figure 4.
Effect of pH and temperature on phenol biodegradation (initial phenol concentration was 800 mg·L−1). (a) Effect of pH on the removal of phenol; (b) effect of temperature on the removal of phenol.
Several Acinetobacter strains have been reported to grow by using phenol as the sole carbon and energy source. However, suspended A. calcoaceticus cells were partially or fully inhibited at high phenol concentrations. For example, free Acinetobacter cells could only degrade phenol at an initial concentration of ≤1000 mg·L−1 [37]. In particular, Adav and Lee noted that A. calcoaceticus isolated by them was fully inhibited at a phenol concentration >1500 mg·L−1 [38]. In our study the inhibition limit of strain A. calcoaceticus PA was as high as the phenol concentration of 2000 mg·L−1.
4. Conclusions
In conclusion, a bacterial strain capable of degrading phenol was isolated from an oil refinery effluent in South China, and it was identified as Acinetobacter calcoaceticus PA based on the 16S rDNA sequence and the phylogenetic analysis. Acinetobacter calcoaceticus PA has the ability to grow in a liquid medium with phenol at different concentrations as the sole carbon and energy source. The strain was able to degrade 91.6% of the initial 800 mg·L−1 phenol and grow at the phenol concentration of as high as 1700 mg·L−1. The optimal growth conditions for phenol degradation of the strain were at 30 °C and pH 8.0. Regarding that native microbial species were more adaptive than non-indigenous microorganisms in polluted environments, their predominance facilitated the bioremediation of the phenol-contaminated environments. Acinetobacter calcoaceticus PA isolated may be used for the bioremediation of the phenol-contaminated environments in the South China regions.
Acknowledgments
This research was supported by the Natural Science Foundation of Guangdong Province, China (S2013040015162, 2015A030313801), Science and Technology Planning Project of Guangdong Province (2013B021000010, 2014A020216046), Science and Technology Planning Project of Maoming (2014039), Doctoral Science Foundation of Guangdong University of Petrochemical Technology (513033) and Open Fund Project of Technology Research Center for Petrochemical Resources Clean Utilization of Guangdong Province (201516B03).
Author Contributions
Zhenghui Liu and Wenyu Xie designed the experiments; Yang Peng performed the experiments on the enrichment and isolation of the target microorganisms; Shusi Liu carried out the study for morphological, physiological and biochemical characteristics of the microorganisms; Zhenghui Liu analyzed the data and wrote the paper; Zesheng Li and Dehao Li critically reviewed and improved the manuscript. All authors evaluated and revised the manuscript writing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Arutchelvan V., Kanakasabai V., Nagarajan S., Muralikrishnan V. Isolation and identification of novel high strength phenol degrading bacterial strains from phenol-formaldehyde resin manufacturing industrial wastewater. J. Hazard. Mater. 2005;127:238–243. doi: 10.1016/j.jhazmat.2005.04.043. [DOI] [PubMed] [Google Scholar]
- 2.Whiteley A.S., Bailey M.J. Bacterial community structure and physiological state within an industrial phenol bioremediation system. Appl. Environ. Microbiol. 2000;66:2400–2407. doi: 10.1128/AEM.66.6.2400-2407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooived M., Heederik D.J.J., Kogevinas M., Boffetta P., Needham L.L., Patterson D.G., Bueno-de-Mesquita H.B. Second follow-up of a dutch cohort occupationally exposed to phenoxy herbicides, chlorophenols, and contaminants. Am. J. Epidemiol. 1998;147:891–899. doi: 10.1093/oxfordjournals.aje.a009543. [DOI] [PubMed] [Google Scholar]
- 4.Environmental Protection Agency (EPA) Sampling and Analysis Procedure for Screening of Industrial Effluents for Priority Pollutants. EPA; Cincinnati, OH, USA: 1977. [Google Scholar]
- 5.Lin S.H., Chuang T.S. Combined treatment of phenolic wastewater by wet air oxidation and activated sludge. Toxicol. Environ. Chem. 1994;44:243–258. doi: 10.1080/02772249409358063. [DOI] [Google Scholar]
- 6.Zilli M., Converti A., Lodi A., Borghi M.D., Ferraiolo G. Phenol removal from waste gases with a biological filter by Pseudomonas putida. Biotechnol. Bioeng. 1993;41:693–699. doi: 10.1002/bit.260410703. [DOI] [PubMed] [Google Scholar]
- 7.Tay S.-L., Moy B.-P., Maszenan A., Tay J.-H. Comparing activated sludge and aerobic granules as microbial inocula for phenol biodegradation. Appl. Microbiol. Biotechnol. 2005;67:708–713. doi: 10.1007/s00253-004-1858-1. [DOI] [PubMed] [Google Scholar]
- 8.Barbosa T.C.P., Luz A.P., Bedin M.L., Gabilan N.H. Effect of ceramic plant effluent on phenol degradation by Acinetobacter calcoaceticus. Int. Biodeter. Biodegr. 1996;37:122. doi: 10.1016/0964-8305(96)84345-9. [DOI] [Google Scholar]
- 9.Paller G., Hommel R.K., Kleber H.-P. Phenol degradation by Acinetobacter calcoaceticus NCIB 8250. J. Basic. Microbiol. 1995;35:325–335. doi: 10.1002/jobm.3620350508. [DOI] [PubMed] [Google Scholar]
- 10.Geng A., Soh A., Lim C., Loke L. Isolation and characterization of a phenol-degrading bacterium from an industrial activated sludge. Appl. Microbiol. Biotechnol. 2006;71:728–735. doi: 10.1007/s00253-005-0199-z. [DOI] [PubMed] [Google Scholar]
- 11.Arutchelvan V., Kanakasabai V., Elangovan R., Nagarajan S., Muralikrishnan V. Kinetics of high strength phenol degradation using Bacillus brevis. J. Hazard. Mater. 2006;129:216–222. doi: 10.1016/j.jhazmat.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 12.Müller C., Petruschka L., Cuypers H., Burchhardt G., Herrmann H. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhIR. J. Bacteriol. 1996;178:2030–2036. doi: 10.1128/jb.178.7.2030-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prieto M.B., Hidalgo A., Serra J.L., Llama M.A.J. Degradation of phenol by Rhodococcus erythropolis UPV-1 immobilized on biolite® in a packed-bed reactor. J. Biotechnol. 2002;97:1–11. doi: 10.1016/S0168-1656(02)00022-6. [DOI] [PubMed] [Google Scholar]
- 14.Tam L.T., Eymann C., Albrecht D., Sietmann R., Schauer F., Hecker M., Antelmann H. Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis. Environ. Microbiol. 2006;8:1408–1427. doi: 10.1111/j.1462-2920.2006.01034.x. [DOI] [PubMed] [Google Scholar]
- 15.Yao R.-S., Sun M., Wang C.-L., Deng S.-S. Degradation of phenolic compounds with hydrogen peroxide catalyzed by enzyme from Serratia marcescens AB 90027. Water Res. 2006;40:3091–3098. doi: 10.1016/j.watres.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Karigar C., Mahesh A., Nagenahalli M., Yun D.J. Phenol degradation by immobilized cells of Arthrobacter citreus. Biodegradation. 2006;17:47–55. doi: 10.1007/s10532-005-3048-y. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Y., Wen J., Bai J., Jia X., Hu Z. Biodegradation of phenol at high initial concentration by Alcaligenes faecalis. J. Hazard. Mater. 2007;147:672–676. doi: 10.1016/j.jhazmat.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y.J., Zhang A.N., Wang X.C. Biodegradation of phenol by using free and immobilized cells of Acinetobacter sp. XA05 and Sphingomonas sp. FG03. Biochem. Eng. J. 2009;44:187–192. doi: 10.1016/j.bej.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Ahmad S.A., Shamaan N.A., Arif N.M., Koon G.B., Shukor M.Y.A., Syed M.A. Enhanced phenol degradation by immobilized Acinetobacter sp. strain AQ5NOL 1. World J. Microbiol. Biotechnol. 2012;28:347–352. doi: 10.1007/s11274-011-0826-z. [DOI] [PubMed] [Google Scholar]
- 20.Maeng J.H., Sakai Y., Tani Y., Kato N. Isolation and characterization of a novel oxygenase that catalyzes the first step of n-alkane oxidation in Acinetobacter sp. strain M-1. J. Bacteriol. 1996;178:3695–3700. doi: 10.1128/jb.178.13.3695-3700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luckarift H.R., Sizemore S.R., Farrington K.E., Fulmer P.A., Biffinger J.C., Nadeau L.J., Johnson G.R. Biodegradation of medium chain hydrocarbons by Acinetobacter venetianus 2AW immobilized to hair-based adsorbent mats. Biotechnol. Progr. 2011;27:1580–1587. doi: 10.1002/btpr.701. [DOI] [PubMed] [Google Scholar]
- 22.Silva Í.S., Santos E.d.C.d., Menezes C.R.d., Faria A.F.D., Franciscon E., Grossman M., Durrant L.R. Bioremediation of a polyaromatic hydrocarbon contaminated soil by native soil microbiota and bioaugmentation with isolated microbial consortia. Bioresource Technol. 2009;100:4669–4675. doi: 10.1016/j.biortech.2009.03.079. [DOI] [PubMed] [Google Scholar]
- 23.Kloos W.E., Tornabene T.G., Schleifer K.H. Isolation and characterization of micrococci from human skin, including two new species: Micrococcus lylae and Micrococcus kristinae. Int. J. Syst. Bacteriol. 1974;24:79–101. doi: 10.1099/00207713-24-1-79. [DOI] [Google Scholar]
- 24.Holt S.G., Kriey N.R., Sneath P.H.A., Staley J.T., Williams S.T. Bergy's Manual of Determinative for Bacteriology. Williams and Wilkins; New York, NY, USA: 1998. [Google Scholar]
- 25.Lanyl B. Classical and rapid identification methods for medically important bacteria. Method. Microbiol. 1987;19:1–67. [Google Scholar]
- 26.Maszenan A.M., Seviour R.J., Patel B.K.C., Rees G.N., McDougall B.M. Amaricoccus gen. nov., a gram-negative coccus occurring in regular packages or tetrads, isolated from activated sludge biomass, and descriptions of Amaricoccus veronensis sp. nov., Amaricoccus tamworthensis sp. nov., Amaricoccus macauensis sp. nov., and Amaricoccus kaplicensis sp. nov. Int. J. Syst. Evol. Microbiol. 1997;47:727–734. doi: 10.1099/00207713-47-3-727. [DOI] [PubMed] [Google Scholar]
- 27.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K., Dudley J., Nei M., Kumar S. Mega4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 30.American Public Health Association (APHA) Standard Methods for the Examination of Water and Wastewater. American Public Health Association, American Water Works Association and Water Environment Federation; Washington, DC, USA: 2005. [Google Scholar]
- 31.Dong X., Hong Q., He L., Jiang X., Li S. Characterization of phenol-degrading bacterial strains isolated from natural soil. Int. Biodeter. Biodegr. 2008;62:257–262. doi: 10.1016/j.ibiod.2008.01.011. [DOI] [Google Scholar]
- 32.Ishii S.i., Koki J., Unno H., Hori K. Two morphological types of cell appendages on a strongly adhesive bacterium, Acinetobacter sp. strain Tol 5. Appl. Environ. Microbiol. 2004;70:5026–5029. doi: 10.1128/AEM.70.8.5026-5029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Y., Chen M., Zhang W., Lin M. Genetic organization of genes encoding phenol hydroxylase, benzoate 1,2-dioxygenase alpha subunit and its regulatory proteins in Acinetobacter calcoaceticus PHEA-2. Curr. Microbiol. 2003;46:235–240. doi: 10.1007/s00284-002-3840-4. [DOI] [PubMed] [Google Scholar]
- 34.Zhan Y., Yan Y., Zhang W., Yu H., Chen M., Lu W., Ping S., Peng Z., Yuan M., Zhou Z., et al. Genome sequence of Acinetobacter calcoaceticus PHEA-2, iolated from industry wastewater. J. Bacteriol. 2011;193:2672–2673. doi: 10.1128/JB.00261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun J.Q., Xu L., Tang Y.Q., Chen F.M., Wu X.L. Simultaneous degradation of phenol and n-hexadecane by Acinetobacter strains. Bioresource Technol. 2012;123:664–668. doi: 10.1016/j.biortech.2012.06.072. [DOI] [PubMed] [Google Scholar]
- 36.Jiang L., Ruan Q., Li R., Li T. Biodegradation of phenol by using free and immobilized cells of Acinetobacter sp. BS8Y. J. Basic Microbiol. 2013;53:224–230. doi: 10.1002/jobm.201100460. [DOI] [PubMed] [Google Scholar]
- 37.Adav S.S., Chen M.Y., Lee D.J., Ren N.Q. Degradation of phenol by Acinetobacter strain isolated from aerobic granules. Chemosphere. 2007;67:1566–1572. doi: 10.1016/j.chemosphere.2006.11.067. [DOI] [PubMed] [Google Scholar]
- 38.Adav S.S., Lee D.J. Single-culture aerobic granules with Acinetobacter calcoaceticus. Appl. Microbiol. Biotechnol. 2008;78:551–557. doi: 10.1007/s00253-007-1325-x. [DOI] [PubMed] [Google Scholar]