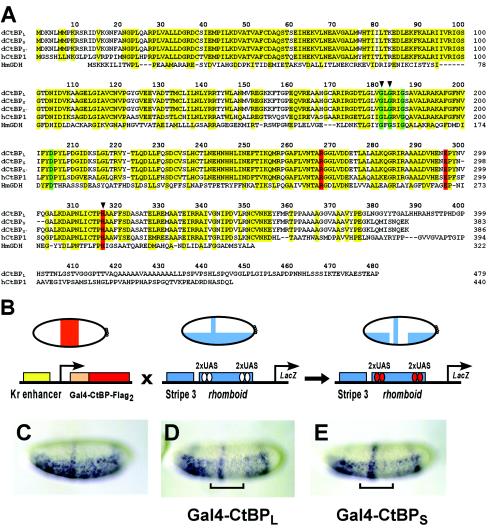
FIG. 1.
CtBP protein sequences and functional assays in the Drosophila embryo. (A) Sequence alignment of CtBP proteins from D. melanogaster (the three dCtBP splice forms are designated L, S, and S′), hCtBP1, and the apoenzyme form of d-glycerate dehydrogenase from Hyphomicrobium methylovorum (HmGDH). Conserved residues in CtBP proteins, as well as those residues conserved in d-glycerate dehydrogenase, are indicated in yellow. Residues involved in NAD binding and the catalytic triad are displayed in green and red, respectively. Arrowheads indicate residues that were altered in the mutant forms of Gal4-CtBPL tested and shown in Fig. 7. (B) In vivo repression assay. Flies expressing Gal4-CtBPL or Gal4-CtBPS under the control of the Krüppel enhancer were crossed with flies containing the integrated eve stripe 3/rhomboid lacZ reporter transgene. Reporter gene expression was visualized in embryos by in situ hybridization with a lacZ antisense RNA probe. (C) Expression pattern of the unrepressed eve stripe 3/rhomboid lacZ reporter gene. (D and E) CtBPL (D) and CtBPS (E) exhibit similar levels of repression activity of the rhomboid enhancer (indicated by brackets). eve stripe 3 expression is unaffected, consistent with short-range repression. In this and subsequent figures, lateral or ventrolateral views of embryos are shown, anterior to left.