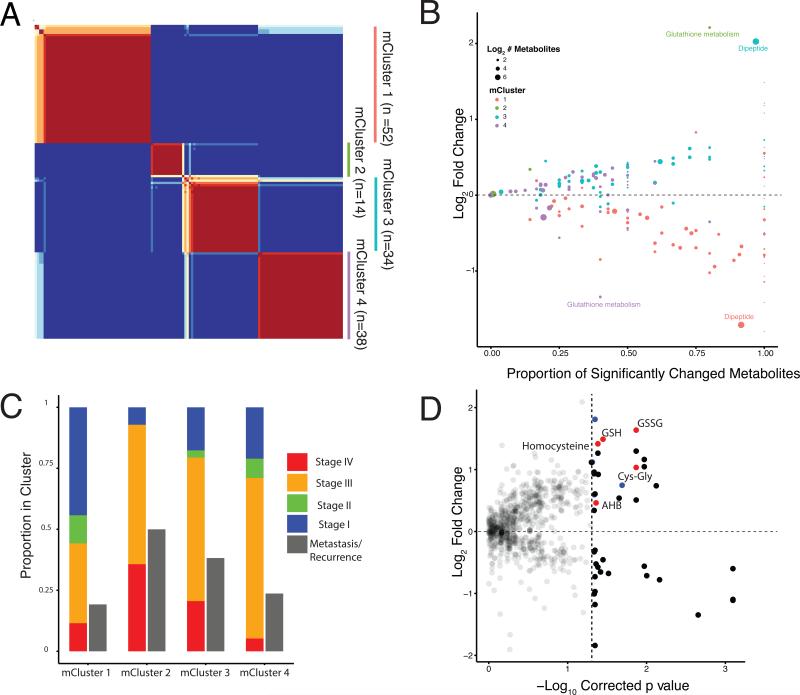
Figure 4. Unsupervised clustering of ccRCC based on metabolite signatures.
(A) Nonnegative matrix factorization (NMF) clustering of metabolomics data. Note that consensus results show consistency for k=4. (B) Mann-Whitney U-tests were used to calculate which metabolites were significantly increased or decreased in each cluster, relative to all other tumors (Benjamini-Hochberg corrected p value < 0.05). X-axis indicates, for a given cluster of patient samples, the proportion of metabolites in a pathway that are significantly changed (both increased and decreased) in a cluster. Y-axis plots the average log2 fold change of these metabolites. mCluser 2 & mCluser 4 are enriched in either increase or decrease of metabolites concerning glutathione metabolism, respectively. mCluster 1 & 3 show large differences in dipeptide levels, relative to other tumor samples. (C) The clinical stages at sample collection and the eventual metastasis of each individual metabolic cluster are presented. mCluster 1 is particularly enriched with Stage 1 tumors (p<0.0001 Chi-Square). (D) Comparison of metabolite abundances in tumors developing metastases versus those not developing metastases at preparation of this report. Red, the metabolites in the glutathione biosynthetic pathway that are increased in tumors that developed metastases. Blue dots correspond to dipeptides. See also Table S5 and S6 and Figure S3.