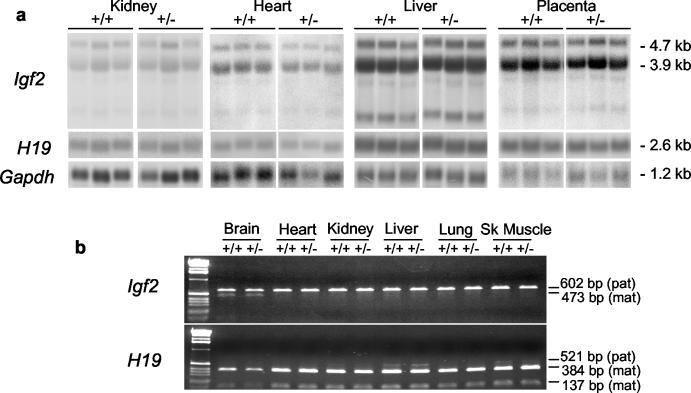
FIG. 3.
Analysis of Igf2 and H19 expression levels and imprinting after paternal transmission of H19ΔK deletion. (a) Heterozygous H19ΔK males were mated to C57BL/6J females, and the expression levels of Igf2 and H19 in the newborn H19ΔK (+/−) offspring and their wild-type (+/+) littermates were analyzed by Northern blot along with E18 H19ΔK and wild-type placentas. RNA was electrophoresed under denaturing conditions and hybridized with probes specific for Igf2, H19, and Gapdh. Six samples of each genotype from each tissue were quantified with a phosphorimager (Fujifilm FLA-3000 and AIDA software) (three representative samples are shown). The total Igf2 and H19 transcripts were normalized against Gapdh levels. A t test was carried out and showed no significant difference in Igf2 and H19 expression levels between wild-type and H19ΔK tissues. (b) Heterozygous H19ΔK males were mated to homozygous SD7 females, and allele-specific expression of Igf2 and H19 was analyzed in tissues from newborn H19ΔK (+/−) offspring and their wild-type (+/+) littermates by RT-PCR. For Igf2, an SD7-specific BsaAI polymorphism was used to distinguish between parental alleles (602-bp band for C57BL/6 [paternal] and 473-bp band for SD7 [maternal]). For H19, an SD7-specific BglI polymorphism was used to distinguish between parental alleles (521-bp band for C57BL/6 [paternal] and 384- and 137-bp bands for SD7 [maternal]). Two samples of each tissue were analyzed. No change in imprinted expression of Igf2 or H19 was detected.