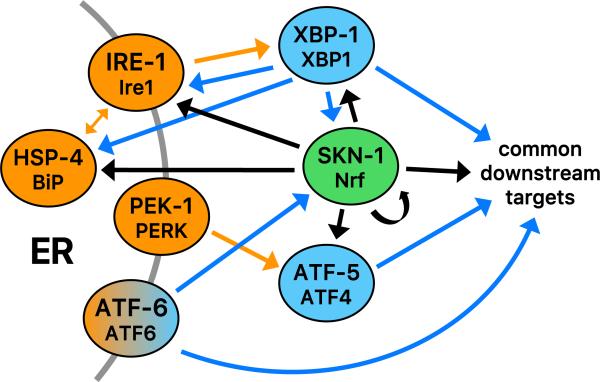
Fig. 4.
Integration of SKN-1 into the UPR. Unfolded proteins that accumulate in the ER (ER stress) are sensed by the transmembrane proteins ATF-6, PEK-1 (PERK), and IRE-1, acting in conjunction with the ER chaperone HSP-4 (BiP) [59,60]. Under ER stress conditions, the transcription factor ATF-6 is released by processing in the Golgi, and the PEK-1 (PERK) kinase inhibits translation by phosphorylating the initiation factor eIF-2α. Lower levels of translation reduce the secretory load, but also result in preferential translation of the transcription factor ATF-5 (ATF4). ER stress also induces IRE-1 to splice the mRNA encoding the transcription factor XBP-1, leading to synthesis of an active XBP-1 form. XBP-1 controls the greatest proportion of the UPR. Genetic, gene expression, and ChIP studies indicate that SKN-1 directly activates transcription of ire-1, xbp-1, hsp-4, atf-5, and itself, and cooperates with the UPR transcription factors to induce expression of downstream UPR target genes [24]. In turn, skn-1 expression is up-regulated by ATF-6 and XBP-1. The UPR encompasses activation of ER function, chaperone, and stress response genes [59] and overlaps with but is distinct from the SKN-1-mediated oxidative stress response [24]. Unfolded protein sensors or chaperones are shown in orange, and canonical UPR transcription factors in blue. Functional interactions in the canonical UPR are shown with orange arrows [59,60]. Direct transcriptional effects of canonical UPR transcription factors and SKN-1 are diagrammed with blue and black arrows, respectively [24].