Table 8.
In Vitro DMPK Profiling of Select 5-Chloropyridin-2-yl Ether Analogues
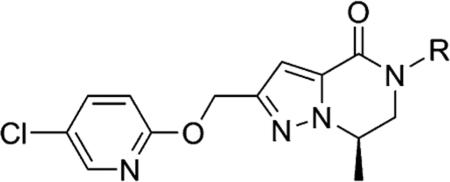
| |||||||
|---|---|---|---|---|---|---|---|
| no. | R | cLogPa | LLEb | mGlu3 IC50 (nM) | fold vs mGlu5 | rat plasma Fuc | rat CLhep (mL/min/kg)d |
| 99 | 2-cyano-5-fluorophenyl | 3.29 | 2.99 | 529 | >56 | 0.045 | 47.7 |
| 100 | pyridin-2-yl | 2.57 | 3.91 | 328 | >91 | 0.051 | 69.5 |
| 101 | pyridin-3-yl | 2.16 | 4.06 | 608 | >49 | 0.118 | 42.0 |
| 106 | 2-fluoropyridin-3-yl | 2.67 | 3.74 | 392 | >76 | 0.083 | 36.9 |
| 107 | 4-fluoropyridin-3-yl | 2.26 | 4.06 | 482 | >62 | 0.085 | 36.0 |
| 108 | 5-fluoropyridin-3-yl | 2.26 | 4.06 | 481 | >62 | 0.092 | 26.6 |
| 109 | 5-cyanopyridin-3-yl | 1.88 | 4.34 | 605 | >49 | 0.078 | 22.8 |
Calculated using Dotmatics Elemental (www.dotmatics.com/products/elemental/).
LLE (ligand-lipophilicity efficiency) = pIC50 – cLogP.
Fu = fraction unbound.
Predicted hepatic clearance based on intrinsic clearance in rat liver microsomes.
