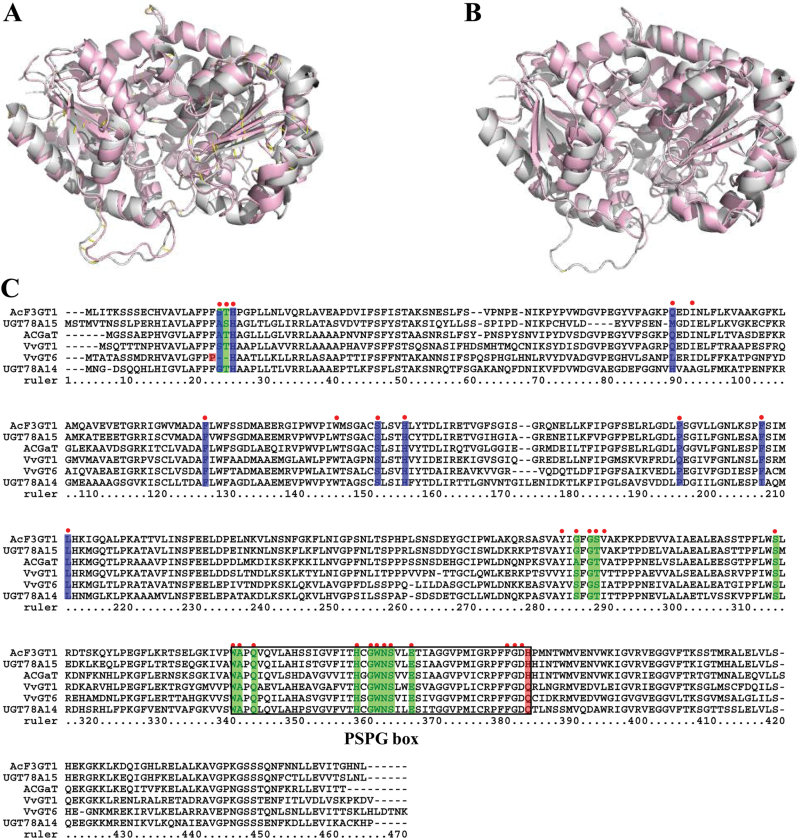
Fig. 4.
Homology modelling and multiple sequence alignment of several UGTproteins at amino acid level. (A, B) Homology models of CsUGT78A14 (A) and CsUGT78A15 (B) were constructed with 2c9z of VvGT1 as the template model. The models of VvGT1 are in grey, and CsUGT78A14 (A) and CsUGT78A15 (B) are shown in light red. (C) Multiple sequence alignment of CsUGT78A14 (KP682360) and CsUGT78A15 (KP682361) with identified UGTs including VvGT1 (AAB81683.1), VvGT6 (BAI22847.1), ACGaT (BAD06514.1), and AcF3GT1 (ADC34700.1). The multiple sequence alignment was performed using a ClustalW program. The amino acid residues for ligands are denoted with red dots, active sites for UDP-sugar donor and active sites for sugar acceptor are, respectively, highlighted in green and blue. The key residues determining the GalT and GlcT activity specificity are highlighted in red. The PSPG box of these aligned sequences is indicated by a black rectangle.