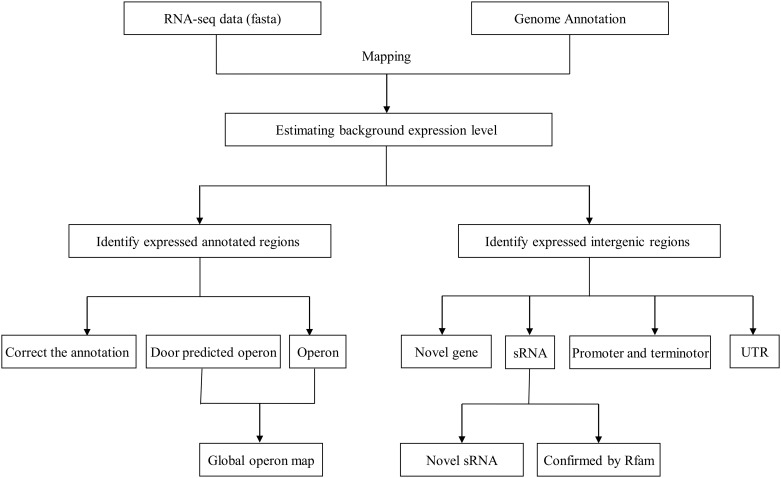
Fig 2. Worlflow of RNA-Seq data analysis.
RNA-seq reads were mapped to the genome using the alignment program BWA. Combining with the existing annotation, expressed annotated regions and expressed intergenic regions are generated by using SAMTools and Perl scripts. Novel protein coding genes, UTRs and sRNAs are identified.