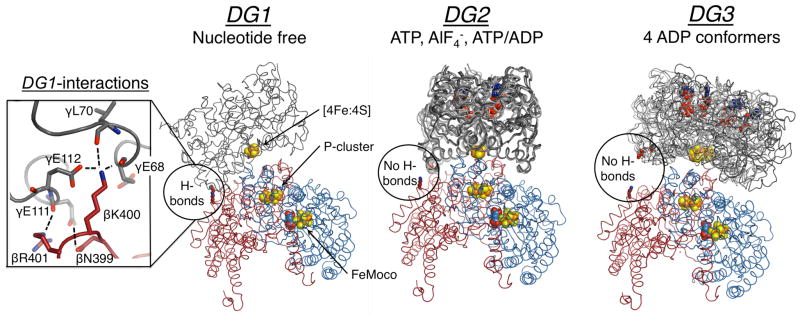
Figure 1.
Structures of the nucleotide free, ATP, and ADP bound nitrogenase, where FeP (γ-subunit) is grey, MoFeP α-subunit is blue, and MoFeP β-subunit is red. The location of the β399–401 surface patch is circled, and the inset on the left shows interprotein interactions in DG1. All known FeP conformations in each docking geometry are depicted. For DG1 the only available structure is shown (PDB ID: 2AFH). For DG2, AMPPCP (a nonhydrolyzable ATP analog) (PDB ID: 2AFK), ADP.AlF4− (PDB ID: 1M34), and ATP/ADP-bound (PDB ID: 4WZA) structures are shown. For DG3, all four ADP-bound FeP conformers (PDB ID: 2AFI) are overlaid. Oxygen, nitrogen, iron, and sulfur are colored red, blue, orange, and yellow, respectively. Hydrogen bonds are marked by dashed lines. Nucleotides and metal clusters are shown as spheres. Only one αβ MoFeP dimer is displayed.