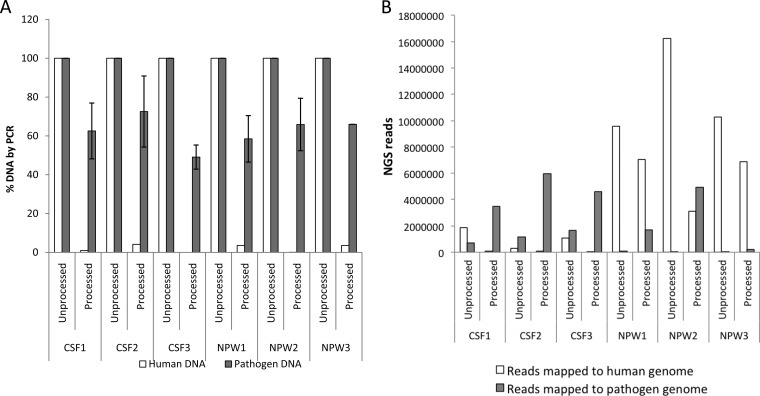
FIG 4.
Analysis of clinical specimens processed with 0.025% saponin and DNase by PCR and NGS. CSF specimens (n = 3) spiked with S. pneumoniae, N. meningitidis, H. influenzae, E. coli, S. agalactiae, HSV2, and adenovirus and NPA specimens (n = 3) spiked with S. pneumoniae, H. influenzae, B. pertussis, HSV2, and adenovirus were treated with saponin to a final concentration of 0.025%, followed by DNase treatment (see Materials and Methods). (A) Nucleic acids extracted from processed or unprocessed samples were analyzed by real-time PCR assays for β-actin DNA for human DNA, and pathogen-specific targets, as described in Table 1. The percentage of DNA was obtained from fold changes calculated from CT values, with respect to CT values for unprocessed samples. Data for all spiked pathogens were averaged; error bars are the standard error of the mean. (B) Nucleic acids extracted from processed or unprocessed samples were analyzed by NGS on Illumina MiSeq. Sequencing libraries were constructed from 1 ng of DNA from specimens using Nextera XT. Raw data were filtered for quality and mapped to human genome and pathogen genomes, as described in the Materials and Methods. Specimens were processed and analyzed independently, as they were available.