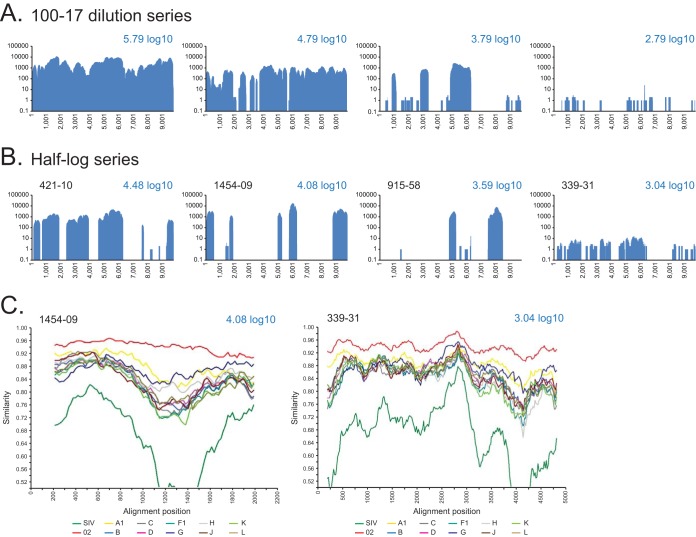
FIG 5.
Coverage plots of HIV-SMART NGS data for CRF02-infected clinical specimens. (A) Serial dilution of 100-17 neat and at 1:10, 1:100, and 1:1,000 with the calculated viral load shown; coverage is based on reads aligned to the 100-17 consensus genome sequence. (B) Patient specimens (ID indicated) with viral loads varying from 3 to 4.5 log10 copies/ml. Coverage is based on reads aligning to the CRF02 reference, GHNJ188. (C) SimPlot analysis was performed with gap-stripped alignments of partial genomes using a 400-bp window, 20-bp step, and Kimura 2-parameter test, with a transition/transversion (T/t) value of 2.0 for specimens 1454-09 (4.08 log10; 2,104-bp alignment) and 339-31 (3.04 log10; 4,923-bp alignment).