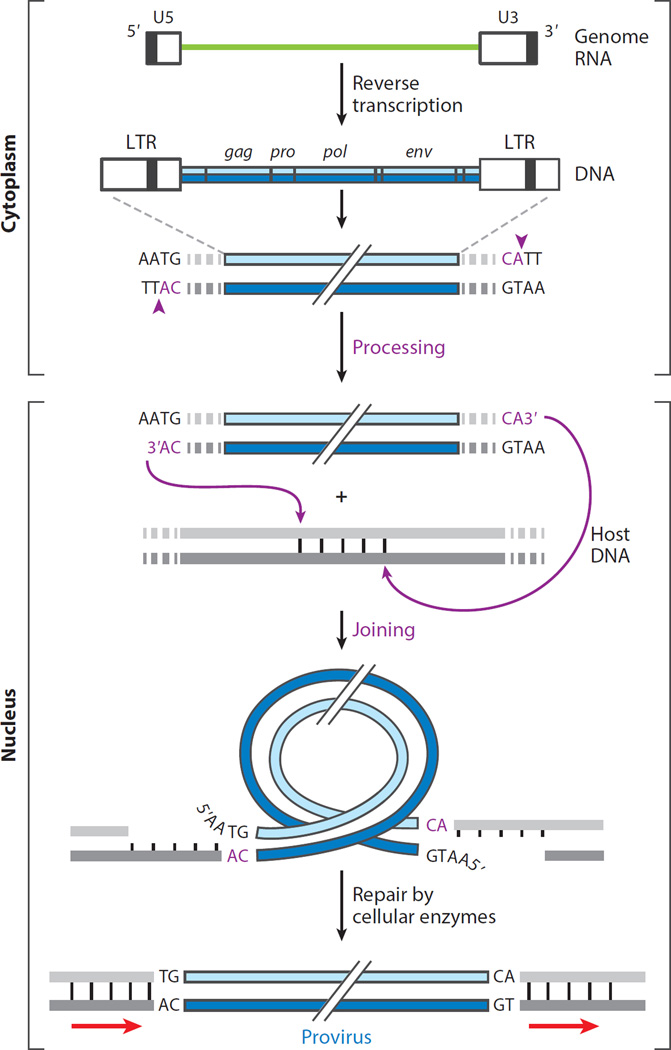
Figure 1.
Steps in the synthesis of retroviral DNA and its integration into host DNA. The viral RNA genome (green line) is reverse transcribed in the cytoplasm of the cell within a subviral nucleoprotein structure (called the reverse transcription complex) to form a duplex DNA containing long terminal repeats (LTRs) of sequences unique to the 5′ (U5) and 3′ (U3) ends of the viral RNA. The organization of the genes common to all retroviruses (gag, pro, pol, and env) is colinear with the RNA genome. Imperfect inverted repeats at the LTR duplex termini are recognized and nicked by cognate integrase (IN) proteins, following a conserved CA dinucleotide at each 3′ end (vertical arrows in inset), producing recessed 3′-OH ends. This first reaction catalyzed by IN is called processing and takes place within a nucleoprotein assembly called a preintegration complex. A tetramer of IN bound to the processed viral DNA (vDNA) ends enters the nucleus where a joining reaction catalyzed by IN connects the 3′-OH ends of the vDNA to staggered phosphates at a target site in the host DNA. The length of the stagger (4–6 bp) is characteristic of the viral IN protein. The conserved CA dinucleotide and steps catalyzed by IN are highlighted in magenta. Removal of the noncomplementary 5′ nucleotides of vDNA and repair of the gaps in host DNA by host enzymes generate a covalently integrated provirus, shorter by 2 bp on either end and flanked by duplications of the target site (arrows).