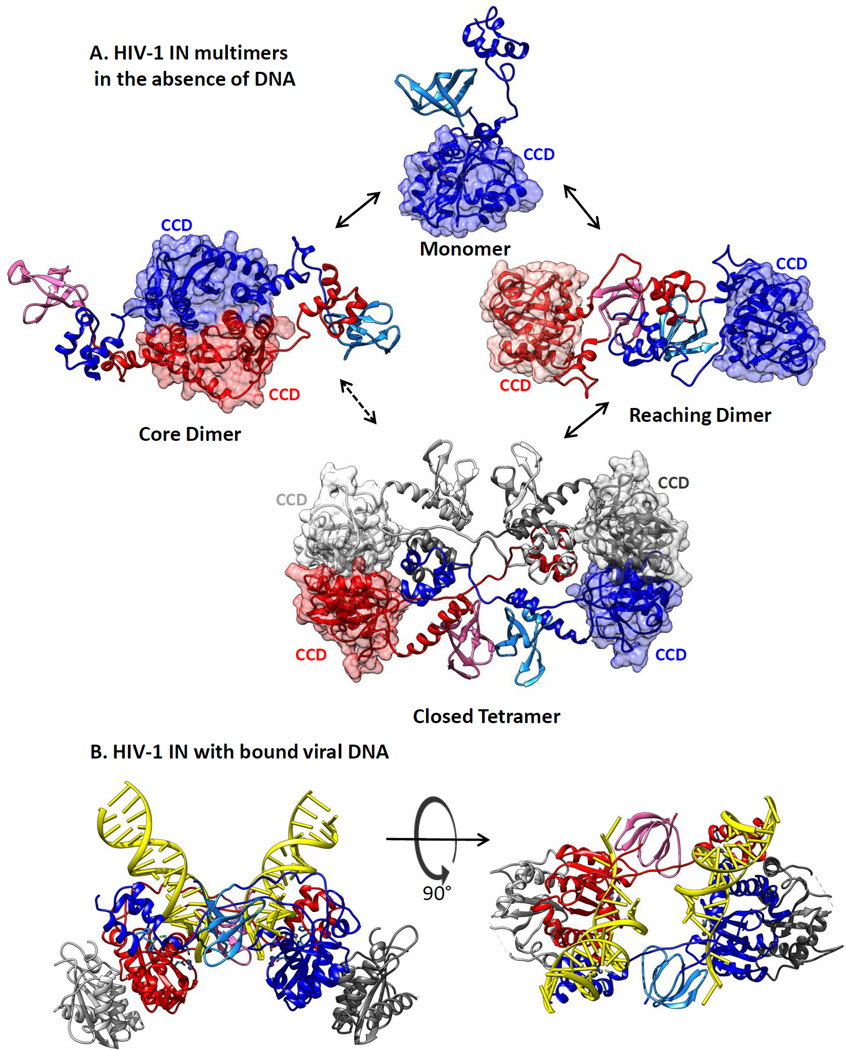
Figure 4.
Models for architectures of full-length human immunodeficiency virus type 1 (HIV-1) apo-integrase (IN) protein in solution. (a) Structures for HIV-1 IN protein in the absence of DNA substrates were derived by HADDOCK data-driven modeling of the HIV-1 IN monomer, dimer, and tetramer in solution, based on small-angle X-ray scattering and protein cross-linking data (93). Catalytic core domains (CCDs) are rendered in muted surface representation to emphasize their locations in the structures. It is not yet known which of these forms is competent for the viral DNA binding that leads to assembly of an HIV-1 intasome. (b) An HIV-1 intasome model (90) with DNA shown in yellow ladder representation. Structures, colored as in Figure 3, are in a ribbon rendering and were generated using Chimera software (133).