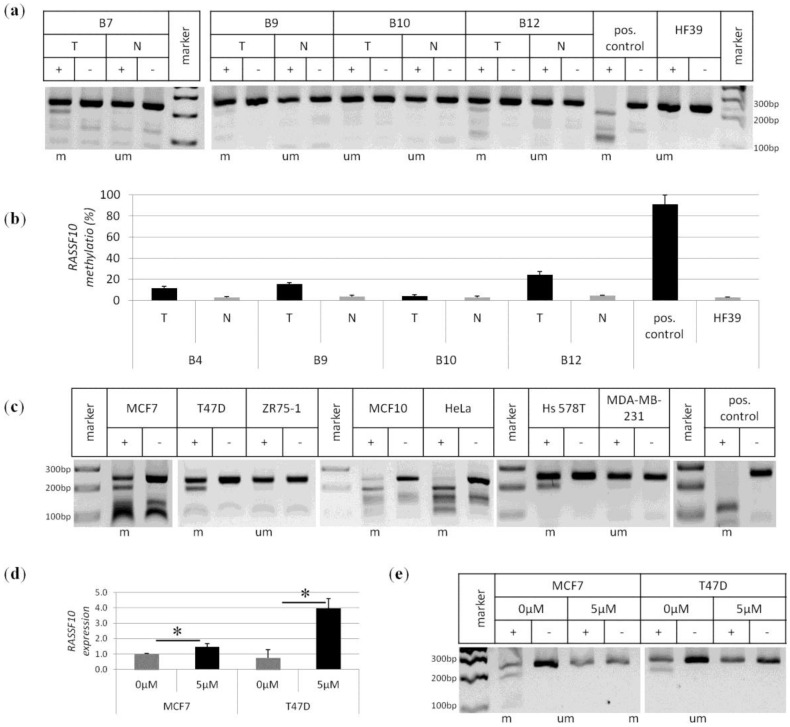
Figure 2.
RASSF10 promoter methylation in primary breast tumours and breast cancer cell lines. (a) RASSF10 COBRA promoter methylation analysis is shown exemplarily for samples of primary breast cancer (T) and normal matching control tissues (N). Positive control shows full methylation of RASSF10 promoter vs. human fibroblasts HF39 as negative control. PCR products for the RASSF10 promoter from bisulfite treated DNA were digested (+) with TaqI or mock digested (−). According digestion products were separated on 2% TBE gel together with 100 bp marker. Methylated samples (m) and unmethylated samples (um) are indicated; (b) RASSF10 promoter methylation was quantified by pyrosequencing for breast tumour samples (T) in comparison to normal matching control samples (N). As methylation controls HeLa (positive control) and human fibroblasts (HF39, negative control) were used. Mean of seven CpGs is shown; (c) RASSF10 promoter methylation of cancer cell lines is shown by COBRA. Breast cancer cell lines MCF7, T74D, MCF10 and Hs578T are methylated to a varying degree, but ZR75-1 and MDA-MB-231 were unmethylated for RASSF10. As positive control cervix carcinoma cell line HeLa and ivm DNA (pos. control) were used; (d and e) Treatment of breast cancer cell lines with demethylating agent 5-Aza-2′-deoxycytidin reverses RASSF10 expression repression due to DNA promoter demethylation. Cell lines MCF7 and T47D were treated with 5 μM Aza or mock on four consecutive days followed by DNA and RNA isolation. RNA was reversely transcribed and RASSF10 reexpression was normalised to b-Actin. Significant reexpression (asterisks) was determined by two-tailed T-Test for MCF7 p < 0.05 and T47D p < 0.005 (d); Isolated DNA was bisulfite treated and RASSF10 promoter demethylation was analysed by COBRA (e).