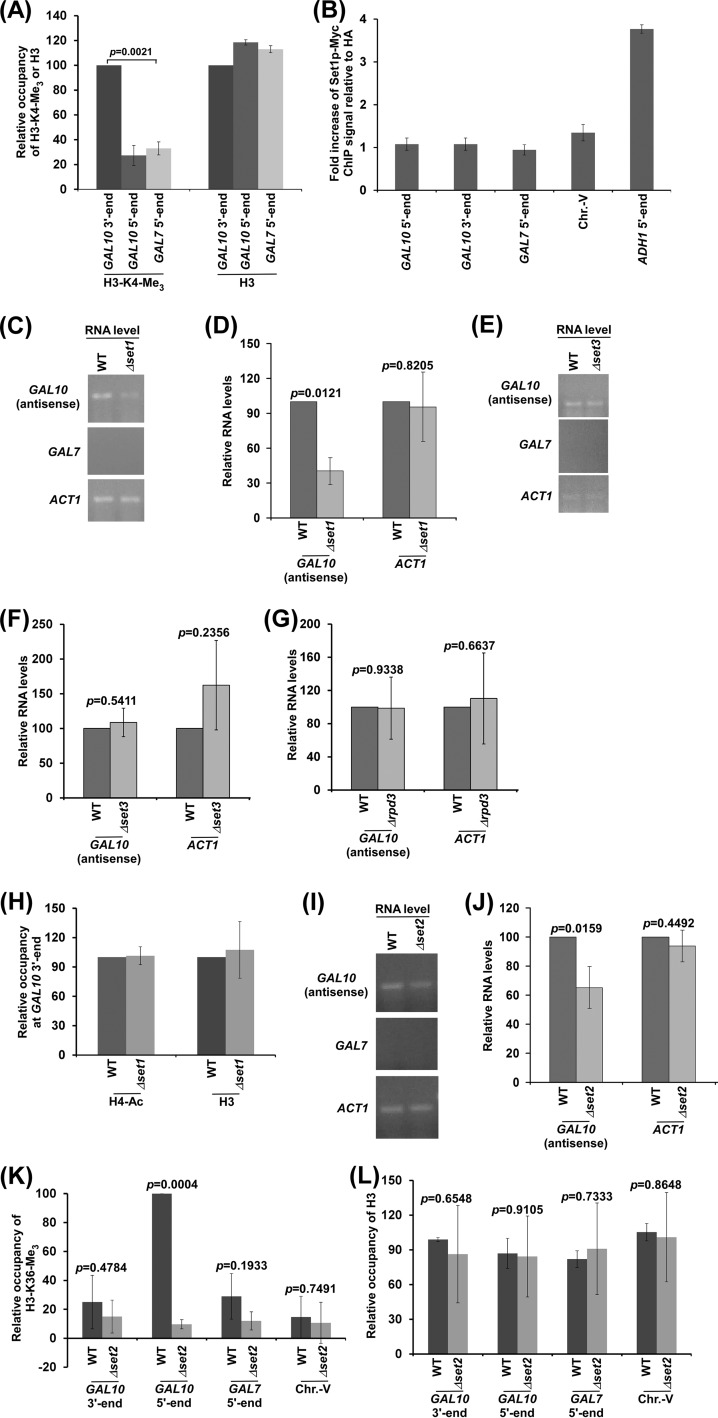
FIG 5.
GAL10 antisense transcription is regulated by histone H3 K4 and K36 methyltransferases. (A) Analysis of histone H3 K4 trimethylation (H3-K4-Me3) and histone H3 levels at the 5′ and 3′ ends of the GAL10 coding sequence and the 5′ end of the GAL7 coding sequence. (B) ChIP analysis of Myc-tagged Set1p at the 5′ and 3′ ends of the GAL10 coding sequence, Chr.-V, and 5′ ends of the ADH1 and GAL7 coding sequence. The fold increase of Set1p ChIP signal relative to HA is plotted. (C and D) RT-PCR analysis of GAL10 antisense RNA in the Δset1 and wild-type strains. The transcription data shown in panel C were plotted in panel D. (E and F) RT-PCR analysis of GAL10 antisense RNA in the Δset3 and wild-type strains. (G) RT-PCR analysis of GAL10 antisense RNA in the Δrpd3 and wild-type strains. (H) ChIP analysis of histone H4 acetylation and histone H3 levels at GAL10 in the wild-type and Δset1 strains. (I and J) RT-PCR analysis of GAL10 antisense RNA in the Δset2 and wild-type strains. (K and L) ChIP analysis of histone H3 K36 trimethylation (H3 K36-Me3) and histone H3 at GAL10, GAL7, and Chr.-V in dextrose-containing growth medium. The maximum ChIP signal was set to 100, and other ChIP signals relative to the maximum ChIP signal were plotted.