Abstract
Ecdysoneless (ECD) is an evolutionarily conserved protein whose germ line deletion is embryonic lethal. Deletion of Ecd in cells causes cell cycle arrest, which is rescued by exogenous ECD, demonstrating a requirement of ECD for normal mammalian cell cycle progression. However, the exact mechanism by which ECD regulates cell cycle is unknown. Here, we demonstrate that ECD protein levels and subcellular localization are invariant during cell cycle progression, suggesting a potential role of posttranslational modifications or protein-protein interactions. Since phosphorylated ECD was recently shown to interact with the PIH1D1 adaptor component of the R2TP cochaperone complex, we examined the requirement of ECD phosphorylation in cell cycle progression. Notably, phosphorylation-deficient ECD mutants that failed to bind to PIH1D1 in vitro fully retained the ability to interact with the R2TP complex and yet exhibited a reduced ability to rescue Ecd-deficient cells from cell cycle arrest. Biochemical analyses demonstrated an additional phosphorylation-independent interaction of ECD with the RUVBL1 component of the R2TP complex, and this interaction is essential for ECD's cell cycle progression function. These studies demonstrate that interaction of ECD with RUVBL1, and its CK2-mediated phosphorylation, independent of its interaction with PIH1D1, are important for its cell cycle regulatory function.
INTRODUCTION
Precisely regulated cell proliferation is essential for embryonic development as well as homeostasis in adult organs and tissues, whereas uncontrolled cell proliferation is a hallmark of cancer (1). A more in-depth understanding of the regulatory controls of cell cycle progression is therefore of great interest.
The Ecd gene was originally inferred from studies of Drosophila melanogaster ecdysoneless (or ecd) mutants that exhibit defective development due to reduced production of the steroid hormone ecdysone (2). Subsequent cloning of drosophila ecd helped identify a cell-autonomous role of ECD protein in cell survival aside from its non-cell-autonomous role in ecdysis (molting) (3). However, the molecular basis of how ECD functions remains unknown (3). The human ECD homologue was initially identified in a screen of human open reading frames that complemented the S. cerevisiae mutants lacking Gcr2 (glycolysis regulation 2) gene, and it rescued the growth defect caused by reduced glycolytic enzyme activity in Gcr2 mutants. The human gene was initially designated HSGT1 (human suppressor of Gcr2) and was suggested to function as a coactivator of glycolytic gene transcription (4). However, ECD protein bears no structural homology to Gcr2, and a true ECD orthologue is absent in S. cerevisiae, suggesting that ECD likely functions by distinct mechanisms.
We identified human ECD in a yeast two-hybrid screen of human mammary epithelial cell cDNA-encoded proteins for novel binding partners of the human papillomavirus 16 (HPV16) E6 oncogene (5). We showed that deletion of Ecd gene in mice causes embryonic lethality, identifying an essential role of ECD during early embryonic development (6). Notably, Cre-mediated conditional deletion of Ecd in Ecdfl/fl mouse embryonic fibroblasts (MEFs) led to a G1/S cell cycle arrest, and this phenotype was rescued by the ectopic expression of human ECD (6), indicating an essential role of ECD in promoting cell cycle progression. We showed that ECD can interact with the retinoblastoma (RB) protein and reduces the repression of RB on E2F transcription factors, providing a novel mechanism by which ECD functions as a positive factor of mammalian cell cycle progression (6). Recently, ECD was shown to play a vital role in pre mRNA splicing by interacting with the pre-mRNA-processing-splicing factor 8 (PRPF8) (7). We and others have shown that ECD shuttles between nucleus and the cytoplasm, with a predominantly cytoplasmic steady-state localization due to rapid nuclear export (7, 8). Consistent with these key cellular roles of ECD, we found that ECD is significantly overexpressed in breast and pancreatic cancers, and its overexpression correlates positively with poor prognostic factors and poor patient survival (9, 10).
A pulldown screen using the phospho-peptide-binding domain of PIH1D1, the adaptor component of the evolutionarily conserved prefoldin-like cochaperone complex R2TP, recently identified ECD as one of the binding partners (11). This interaction was shown to require dual phosphorylation of Ser-505 and Ser-518 on ECD (11), suggesting that ECD phosphorylation may mediate its interaction with the R2TP complex. To date, this interaction has not been demonstrated in the context of endogenous ECD nor has a functional role of this interaction been determined. The core R2TP complex is composed of four proteins: PIH1D1, RPAP3, RUVBL1, and RUVBL2 (each with a number of other names) (12). The R2TP complex is involved in the assembly of multisubunit complexes, including the small nucleolar ribonucleoproteins, RNA polymerase II, and phosphatidylinositol 3-kinase-related kinases and their complexes (13–15). As such, the R2TP complex is involved in a number of essential cellular processes. The closely related RUVBL1 and RUVBL2 proteins are AAA+ (ATPases associated with diverse cellular activities) that are essential for R2TP function (16). Recent studies have shown that RUVBL1 (Pontin) plays an important role in cell cycle regulation (17, 18). Germ line deletion of Ruvbl1 was shown to be early embryonic lethal (18, 19). Depletion of RUVBL1 in AML1-ETO fusion oncogene-expressing leukemic cells was shown to cause cell cycle arrest (17) and Cre-mediated deletion of Ruvbl1 in Ruvbl1fl/fl cells also led to G1/S cell cycle arrest (18). The apparent similarities in the embryonic lethality and cell cycle arrest phenotypes imparted by the loss of ECD or RUVBL1 expression suggested the likelihood that the recently described interaction with the R2TP complex (11) may underlie the functional requirement of ECD in cell cycle progression.
In this study, we extensively analyzed the mechanism of ECD-R2TP interaction and how disabling this interaction by mutations in ECD affects the latter's role in cell cycle progression. We demonstrate that ECD levels and localization do not vary during cell cycle progression. We show that casein kinase 2 (CK2) phosphorylates ECD in cells at 6 major sites and a mutant ECD (6S/A) disabled for CK2-mediated phosphorylation exhibits reduced ability to rescue the cell cycle arrest caused by Ecd gene deletion. Notably, whereas ECD can interact with PIH1D1, loss of this interaction by mutating CK2 phosphorylation sites did not impact the ECD-R2TP association in cells. We identified a novel interaction of ECD with RUVBL1, independent of ECD's interaction with PIH1D1, which we show to be essential for ECD's cell cycle progression function. Notably, a phosphomimetic mutant (6S/D) of ECD failed to bind PIH1D1 and was incompetent at rescuing the cell cycle arrest caused by Ecd gene deletion, suggesting a potential accessory role for PIH1D1-ECD interaction. Taken together, our results demonstrate that although CK2-mediated phosphorylation of ECD is important for its role in cell cycle progression, ECD's interaction with PIH1D1 is dispensable, suggesting that the novel RUVBL1-ECD interaction that we identified is particularly critical for ECD's function in cell cycle.
MATERIALS AND METHODS
Reagents.
λ protein phosphatase (catalog no. P9614) was purchased from Sigma-Aldrich USA, and the treatment was given according to the manufacturer's instructions. 12.5% SuperSep Phos-tag (50 μmol/liter) was purchased from Wako Laboratory Chemicals (catalog no. 195-16391). Electrophoresis was performed according to the manufacturer's protocol. PreScission protease was purchased from GE Healthcare Life Sciences.
Cell cultures.
HEK-293T, MEFs, and T98G glioblastoma cell lines were grown in DMEM (Life Technologies, Grand Island, NY) supplemented with 10% fetal calf serum. Immortal mammary epithelial cell line 76NTERT was cultured in DFCI-1 medium, as described previously (20). U2OS cell line was cultured in α-minimum essential medium (α-MEM). The CK2 inhibitor TBB (4,5,6,7-tetrabromo-2-azabenzimidazole) was dissolved in dimethyl sulfoxide and used at 50 μM.
Plasmid constructs, site-directed mutagenesis, and transfection.
Generation of the pMSCV-puro (Clontech)-based expression constructs for FLAG-ECD, and its truncated versions has been described previously (6, 8). The pMSCV-puro construct expressing ECD with deletion of amino acids 499 to 527 was generated using a thee-fragment ligation into BglII and HpaI sites. C-terminal His6-tagged ECD truncations (1 to 567, 1 to 534, and 1 to 432) were generated through PCR amplification and cloning into XbaI and SalI sites of pET-28b+ vector (Invitrogen), and recombinant proteins were purified after expression in Escherichia coli BL21(DE3) strain using a nickel affinity column (GE Healthcare). C-terminal His6-tagged full-length ECD was generated by cloning ECD coding sequence into SalI and NotI sites of the pFastBac1 vector (Invitrogen), expressed in Sf21 insect cells, and purified by using a nickel affinity column. The PIH1D1-specific and control siRNAs catalog no. Sc-97385; Santa Cruz) were transfected into subconfluent cells using DharmaFECT1 transfection reagent (Thermo Scientific). Green fluorescent protein (GFP)-tagged full-length or truncated ECD expression constructs in the pGEn2 vector were generated by replacing the ST6-Gal1 insert in the ST6-Gal1-pGEn2 construct (ST6GAL1-pXLG-NtermTCMhisStrep-DEST) (21) with PCR-amplified ECD coding sequences at the EcoRI and HindIII sites by infusion cloning kit (Clontech). The primer sequences used for cloning are indicated in Table S2 in the supplemental material. Human PIH1D1 cDNA sequences clone SC321317; Origene) were subcloned into BamHI and XhoI sites of pGEX-6p-1 for expression as a glutathione S-transferase (GST) fusion protein in E. coli BL21. A recombinant GST-PERK kinase domain was expressed in BL21(DE3) cells purified as a GST fusion protein.
Point mutants of ECD were generated using a PCR-based commercial kit (GENEART site-directed mutagenesis system; Invitrogen), according to the manufacturer's instructions, cloned into the pET28b+ vector for His-tagged recombinant protein expression, and purified by using a nickel affinity column. The PCR primer sequences are listed in Table S2 in the supplemental material. All constructs were verified by sequencing.
DNA constructs were transfected in HEK-293T cells using the X-tremeGENE transfection reagent (Roche). Retroviral infection was carried out as described previously (6).
Flow cytometry for cell cycle analysis and biochemical fractionation.
76NTERT cells were plated at 5 × 105 cells per 100-mm dish for 12 h, subjected to growth factor deprivation by culturing in growth factor-free DFCI-3 medium for 72 h (20) and released from synchrony using growth factor-containing DFCI-1 medium (20). Half of the cells were fixed for fluorescence-activated cell sorter (FACS) analysis after fixation in chilled 70% ethanol and staining with propidium iodide; the remaining cells were used for Western blotting. G2/M-to-G1 progression in MEFs was similarly assessed using FACS analysis after nocodazole (100 ng/ml)-dependent arrest in the early G2/M phase of cell cycle (22). Nuclear and cytoplasmic fractions were prepared from cells at various times points during cell cycle progression using the NE-PER kit (Thermo Scientific, catalog no. 78833). Ecdflox/flox MEFs were infected with adeno-Cre-GFP or control adeno-GFP to assess the mitotic index. Cells were collected and fixed as described above. The cell pellet was resuspended in 100 μl of phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and 0.25 μg of phospho-H3-S10 (catalog no. ab14955; Abcam) and then incubated for 1 h at room temperature. Cells were washed in 150 μl of PBS, resuspended in Alexa Fluor 647 (catalog no. A212235; Life Technologies)-conjugated goat anti-mouse antibody diluted at a ratio of 1:300 in 100 μl of PBS containing 1% BSA, and incubated at room temperature in the dark for 30 min, followed by FACS analysis. The median fluorescence intensity (MFI) of GFP-positive cells at 488- and 633-nm wavelengths was recorded as an indicator of mitosis in the control and Ecd-null cells.
Immunoblotting and IP.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris [pH 7.2], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]), and the protein concentration was measured using the bicinchoninic acid (BCA) protein assay reagent (Pierce). Immunoblotting was performed with primary antibodies against ECD (9), RB (catalog no. 554136; Pharmingen), anti-phospho-Ser (05-1000; Millipore), anti-phospho-Thr (AB1607; Millipore), PIH1D1 (sc-101000 or sc-390810; Santa Cruz), RUVBL1 (12300S [Cell Signaling] or SAB4200194 [Sigma]) RUVBL2 (ab36569; Abcam), RPAP3 (HPA038311; Sigma), PARP (sc-8007; Santa Cruz), histone H3 (06-755; Millipore), PRPF8 (ab137694; Abcam), β-actin (A5441; Sigma), or α-tubulin (T6199; Sigma), as indicated. For immunoprecipitations (IPs), the cells were lysed in NP-40 lysis buffer (20 mM Tris-HCl [pH 7.5], 200 mM NaCl, 0.5% Nonidet P-40 [NP-40], 1 mM NaF, 0.1 mM Na3VO4, and protease inhibitor mixture [Roche Applied Science]) and then immunoprecipitated with 3 μg of antibodies against ECD or 35 μl of Ezview red anti-FLAG M2 affinity gel (Sigma) for 2 h to overnight at 4°C. The immune complexes were captured with protein A/G-agarose (sc-2003; Santa Cruz Biotechnology). To elute FLAG-tagged proteins from anti-FLAG beads before analysis, immune complexes were incubated with 150 ng of 3× FLAG peptide (Sigma)/μl for 15 min at room temperature, and the supernatants were collected for SDS-PAGE. For PIH1D1 interaction with ECD, the cell lysates were prepared in CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} lysis buffer (0.3% CHAPS, 0.20 mM Tris-HCl [pH 7.4], 120 mM NaCl, 10% glycerol, 5 mM EDTA) supplemented with protease and phosphatase inhibitor (Roche). RUVBL1 immunoprecipitation to assess association with ECD was carried out using a monoclonal anti-RUVBL1 antibody (catalog no. SAB4200194-200UL [Sigma]; 2 μg). Immunoprecipitated RUVBL1 (close to IgG heavy chain) was detected by Western blotting with an anti-RUVBL1 antibody (catalog no. 12300s; Cell Signaling) that was conjugated to horseradish peroxidase (HRP) using the Lightning-Link HRP conjugation kit (Novus Biologicals).
In vitro kinase assay.
A total of 500 ng of purified recombinant ECD proteins or its mutants was incubated with 0.2 mM ATP, 1 μCi of [γ-32P]ATP (Perkin-Elmer), and 0.2 μl (10 U) of human recombinant CK2 (NEB, Beverly, MA) at 30°C for 30 min or as indicated. The reaction was stopped by adding SDS-PAGE sample buffer. The 32P-labeled proteins were detected by autoradiography after SDS-PAGE and then transfer to polyvinylidene difluoride (PVDF) membranes (Millipore). Once the radioactive signals had decayed, the membranes were blotted with anti-p-Ser antibodies. Ten nanograms of recombinant GST-PERK kinase domain was autophosphorylated in kinase assay buffer (50 mM HEPES [pH 8.0], 10 mM MgCl2, 2.5 mM EGTA) supplemented with 20 μM cold ATP (NEB). Next, 0.5 ng was loaded onto SDS-PAGE gels and subjected to Western blotting with anti-p-Ser and anti-p-Thr antibodies.
32P metabolic labeling and immunoprecipitation.
Exponentially growing or serum-deprived T98G cells were washed with phosphate-free Dulbecco modified Eagle medium (DMEM) supplemented with 10% dialyzed fetal bovine serum and incubated in the same medium for 1 h before adding 0.1 mCi of [32P]orthophosphate (NEN) per 10-cm plate. The cells were labeled for 4 h at 37°C (or for 2, 5, or 16 h for cell cycle analyses), rinsed once in ice-cold PBS, and lysed in ice-cold lysis buffer (250 mM NaCl, 1% NP-40, 20 mM Tris-HCl [pH 7.4], 1 mM EDTA, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 1 μg of pepstatin/ml, 2.5 μg of antipain/ml, 1 μg of chymostatin/ml, 1 mM Na3VO4, 10 mM NaF, 1 mM sodium molybdate, 0.5 mM phenylmethylsulfonyl fluoride). Labeled ECD was immunoprecipitated with affinity-purified mouse anti-Ecd monoclonal antibody or anti-FLAG beads overnight at 4°C, and Protein G Plus/Protein A-agarose beads were added for 1 h. The beads were washed thee times with ice-cold wash buffer (150 mM NaCl, 1% NP-40, 20 mM Tris-HCl [pH 7.4], 1 mM EDTA, and protease inhibitors). The immunoprecipitated proteins were resolved on 7.5% SDS-polyacrylamide gels, transferred to PVDF membranes, and visualized by autoradiography.
In vitro binding assays.
GST-or His-tagged protein pulldowns were performed as described previously (6). FLAG-tagged wild-type (WT) ECD or its mutants (3S/A and 6S/A) were expressed by transient transfection in 293T cells and lysed in CHAPS lysis buffer as described above. Then, 1,000 μg of lysate protein was incubated with 2 μg of bead-bound purified GST-PIH1D1 for 2 h at room temperature, followed by five washes, and then the bound proteins were detected by Western blotting with anti-FLAG antibody. Membranes were stained with Ponceau S to visualize the GST fusion proteins. In vitro tandem affinity purification (TAP) was performed as described previously (23) using purified recombinant ECD with a C-terminal FLAG tag.
Cell proliferation and colony formation assays.
The cell proliferation was analyzed as described previously (6). Briefly, Ecdflox/flox MEFs were infected with adenoviruses encoding GFP-Cre or GFP (control) (University of Iowa Gene Transfer Vector Core) and plated at 104 cells/well in six-well plates, followed by counting of cells at the indicated time points. For assessing colony formation ability, infected cells were plated at 5,000 or 1,000 per well in six-well plates for 10 days, the colonies were stained with crystal violet (0.5% crystal violet in 25% methanol) and solubilized in 10% acetic acid, and then the extent of colony formation was measured by determining the absorbance at 590 nm.
Statistical analysis.
A generalized estimating equation method was used to assess the differences among cell types accounting for the correlated measurement within a sample. Comparisons between the WT and other cell types at a given time were made with a simulation correction (random sampling from a probability distribution). Some results were analyzed using a paired two-tailed Student t test. P values of ≤0.05 were considered statistically significant.
RESULTS
ECD levels and localization do not change during cell cycle progression.
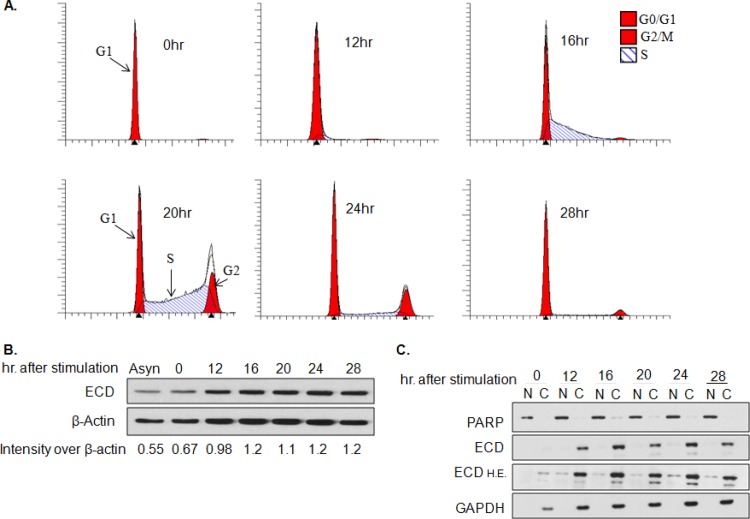
Given the requirement of ECD for cell cycle progression and its direct association with RB (6), we assessed whether ECD levels or localization are altered during cell cycle progression. For this purpose, an immortal mammary epithelial cell line, 76NTERT, was arrested in the G1 cell cycle phase by growth factor deprivation, and the cells were then allowed to proceed synchronously though cell cycle phases by culture in regular growth factor-containing medium. FACS analyses showed that a majority of growth factor-deprived cells were growth arrested, with 98% cells in the G1 phase, and only 0.75% cells in the S phase and 1.25% cells in the G2/M phase (Fig. 1A). Western blotting of lysates showed no significant differences in the levels of ECD protein in cells at various times during cell cycle progression (Fig. 1B). Analysis of nuclear and cytoplasmic fractions prepared at various times during cell cycle progression showed that ECD localizes primarily in the cytoplasm (Fig. 1C), which is consistent with its rapid nuclear export, as previously reported (8). Overall, our results indicate that ECD levels and its subcellular localization do not change significantly during cell cycle progression.
FIG 1.
ECD localization and expression do not change during cell cycle progression. 76NTERT cells were cell cycle arrested by culturing in growth factor-free DFCI-3 medium for 72 h and then switched to growth factor-containing DFCI-1 medium to initiate cell cycle progression. (A) Cells were fixed in 70% ethanol at the indicated time points, stained with propidium iodide, and subjected to FACS analysis. (B) Lysates were collected at the indicated time points and subjected to Western blotting for ECD or β-actin. ImageJ software was used to quantify ECD signals at various time points during cell cycle progression and expressed as relative to β-actin signals. (C) Nuclear (N) and cytoplasmic (C) fractions were isolated from cells at various time points during cell cycle progression and subjected to Western blot analysis with anti-ECD antibody. PARP and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) served as positive controls for nuclear and cytoplasmic proteins, respectively. All experiments were carried out in triplicate. H.E., higher exposure.
ECD is phosphorylated on serine residues, but overall phosphorylation does not change during cell cycle progression.
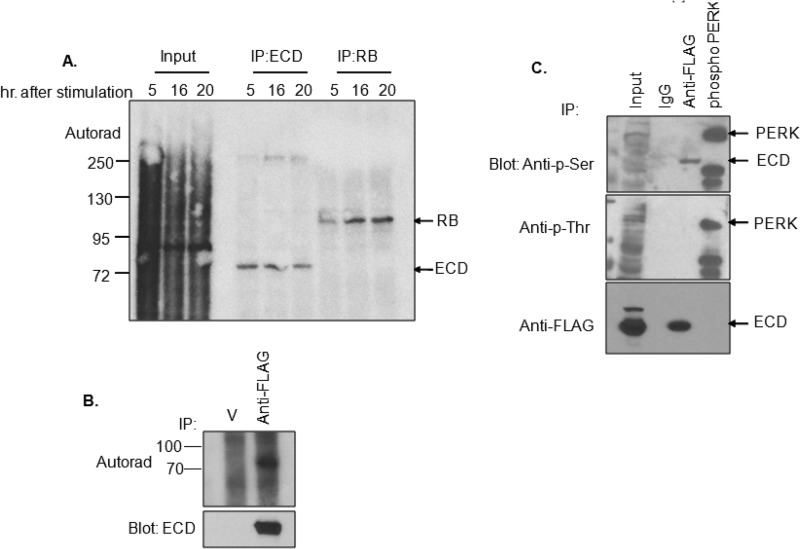
Given the known roles of phosphorylation in regulating the cell cycle machinery (24), we sought to determine whether ECD is a phosphoprotein and whether its phosphorylation varies with cell cycle progression. For these analyses, T98G cells (a human brain glioblastoma cell line that expresses a wild-type RB) (25) were cultured in low-serum medium for 48 h to induce growth arrest and then allowed to progress though cell cycle by adding serum-containing medium with 32P-labeled sodium orthophosphate. Autoradiography of anti-ECD immunoprecipitates showed that ECD is indeed a phosphoprotein; however, the levels of phosphorylation were comparable at various time points during cell cycle progression (Fig. 2A). As a control, RB showed an expected cell cycle-related increase in phosphorylation at the 16- and 20-h time points (Fig. 2A). Further analyses using anti-FLAG immunoprecipitations (IPs) from [32P]orthophosphate-labeled cells expressing an exogenous FLAG-tagged ECD protein confirmed the phosphorylation of ECD in cells (Fig. 2B). These analyses demonstrate that ECD is a phosphoprotein; however, the phosphorylation levels do not change during cell cycle progression.
FIG 2.
ECD is phosphorylated, predominantly on serine residues. (A) T98G cells were serum-deprived for 48 h, the last 4 h in phosphate-free medium, and then cultured for indicated times in complete DMEM containing 100 μCi of sodium [32P]orthophosphate per 10-cm plate. ECD or RB (used as a positive control) were immunoprecipitated, resolved by SDS-PAGE, transferred to a PVDF membrane, and subjected to autoradiography to detect the phosphorylation signal. (B) T98G cells transiently transfected to express FLAG-tagged ECD were cultured for 6 h in phosphate-free DMEM and then metabolically 32P labeled, as described above. Anti-FLAG IPs were visualized by autoradiography. (C) Anti-FLAG IPs of T98G cells transfected with FLAG-tagged ECD were Western blotted with anti-p-Ser or anti-p-Thr antibodies. In vitro-phosphorylated GST-PERK kinase domain was used as a positive control for serine and threonine phosphorylation. The extra band observed in GST-PERK lane is likely a cleavage product of PERK. Arrows point to bands of interest.
It was reported that a peptide sequence derived from ECD was phosphorylated by casein kinase 2 (CK2) in vitro (11). To assess whether the phosphorylation of ECD corresponds to phosphoserine (p-Ser) or phosphothreonine (p-Thr) residues, anti-FLAG immunoprecipitates of T98G cells expressing a FLAG-tagged ECD were blotted with anti-p-Ser or anti-p-Thr antibodies. A recombinant GST-PERK kinase domain, known to undergo autophosphorylation on serine and threonine residues during an in vitro kinase reaction (26), was used as a positive control for serine and threonine phosphorylation. Indeed, both p-Ser and p-Thr signals were detected by blotting of autophosphorylated GST-PERK kinase domain (Fig. 2C). Although no signals were detected with anti-p-Thr antibody blotting of anti-ECD immunoprecipitation, even after long exposures, a specific band was observed with the anti-p-Ser antibody (Fig. 2C). These results suggested that cellular ECD is predominantly phosphorylated on serine residues.
CK2-mediated phosphorylation of ECD is important for its cell cycle regulation function.
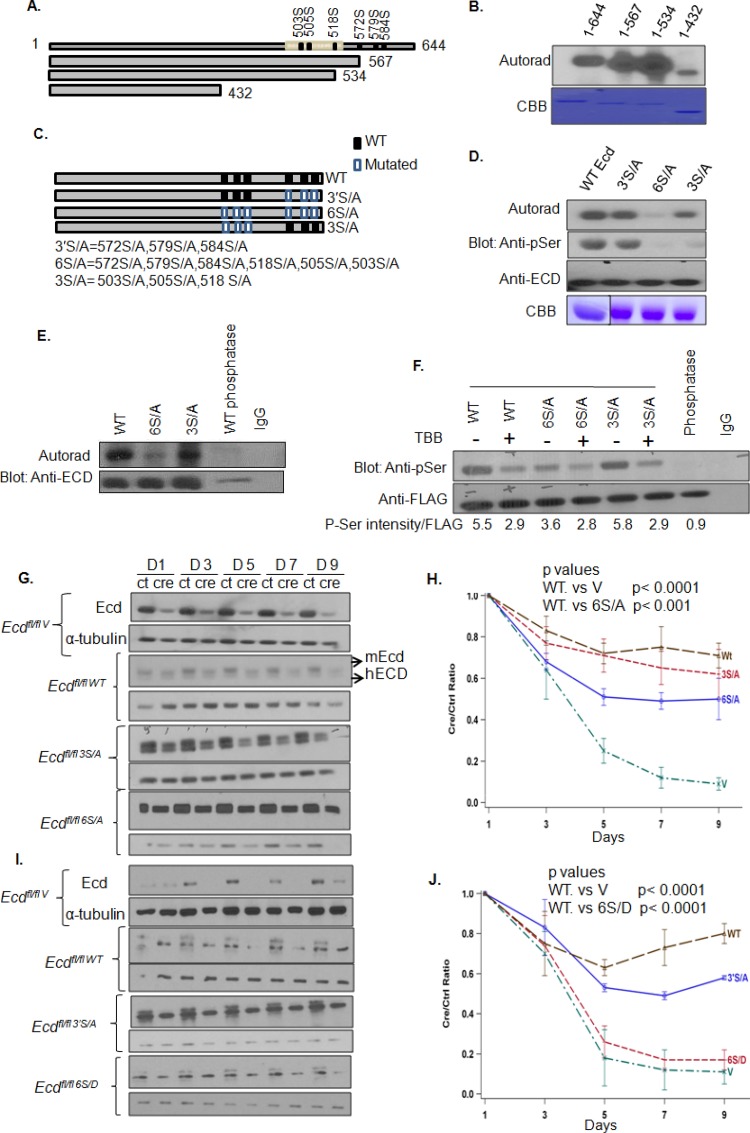
In view of our results presented above, and a recent study that used an array of spotted peptides to identify CK2 phosphorylation of an ECD peptide on Ser-505 and Ser-518 (11), we performed a detailed analysis of potential phosphorylation sites on ECD using the publically available KinasePhos 2.0 tool (http://kinasephos2.mbc.nctu.edu.tw/). This analysis identified multiple sites on ECD that could be phosphorylated by various Ser/Thr kinases. Among these, CK2 was predicted to preferentially phosphorylate multiple serine residues, and this was of obvious interest in view of our results that cellular ECD is primarily phosphorylated on Ser residues (Fig. 2C). The potential CK2 phosphorylation sites near the C terminus, including Ser-505 and Ser-518 reported in the peptide array screen (11), were predicted with the highest confidence (Fig. 3A). To directly assess whether ECD is a CK2 substrate, we performed an in vitro kinase assay with purified CK2 and recombinant full-length ECD protein or its C-terminal truncated versions. Phosphorylation was observed with full-length ECD (residues 1 to 644) and its fragments encompassing residues 1 to 567 or 1 to 534, whereas substantially less phosphorylation was observed with the ECD 1-432 fragment (Fig. 3A and B). These results indicated that ECD was indeed a substrate for CK2 in vitro and that CK2-dependent phosphorylation occurs predominantly within the C-terminal region of ECD.
FIG 3.
Phosphorylation of ECD is important for its ability to rescue cell cycle arrest in Ecd-null MEFs. (A) Schematic of ECD protein, its C-terminal deleted constructs, and CK2 phosphorylation sites predicted by the KinasePhos 0.2 tool (http://kinasephos2.mbc.nctu.edu.tw/). (B) ECD is predominantly phosphorylated near its C terminus. In vitro kinase reactions of full-length (aa 1 to 644) ECD and various C-terminal deletion fragments (aa 1 to 567, 1 to 534, and 1 to 432) with human recombinant CK2 were separated by SDS-PAGE, transferred to PVDF membranes, and subjected to autoradiography to detect 32P signals. The purity of proteins was assessed by Coomassie brilliant blue staining (CBB). (C) Schematic representation of various point mutants. Black rectangles, WT Ser residues; white rectangles, mutant Ala residues. (D) CK2 phosphorylates ECD at six sites. His-tagged wild-type ECD or its point mutants were purified by nickel affinity purification and subjected to an in vitro kinase assay, as described above. 32P labeling was detected using autoradiography, and the filters were subsequently subjected to Western blotting with anti-p-Ser antibody and reprobed with anti-ECD antibody for equal loading. The purity of recombinant proteins was assessed by CBB staining. (E) CK2-dependent phosphorylation of ECD at multiple residues in cultured cells. T98G cells expressing FLAG-tagged ECD or its phosphorylation site mutants 3S/A or 6S/A were metabolically labeled with 32P, as described above. FLAG-tagged ECD and its mutants were immunoprecipitated and subjected to autoradiography. IP of cells expressing WT ECD subjected to phosphatase treatment is shown. The blot was reprobed with anti-ECD antibody for equal IP loading. (F) Inhibition of CK2 reduces ECD phosphorylation. T98G cells expressing FLAG-tagged ECD or its phosphorylation site mutants 3S/A and 6S/A were treated with CK2 inhibitor TBB (50 μM) for 4 h, subjected to anti-FLAG IP, and Western blotted with anti-p-Ser or anti-FLAG antibodies. The intensity of anti-p-Ser signals was quantified using ImageJ software and normalized relative to FLAG-ECD signals. (G) Phosphorylation of ECD at six CK2 sites is important for its cell cycle progression function. Ecdflox/flox MEFs stably expressing vector control, wild-type human ECD, or its indicated mutants were infected with control (ctrl) adeno-GFP or adeno-GFP-Cre (cre) viruses for the indicated times, and lysates were analyzed by anti-ECD and α-tubulin (loading control). Note that human ECD-reconstituted cells express both endogenous mouse (mEcd higher band) and ectopic hECD or its mutants (hECD, lower band). (H to J) Analysis of hECD or its mutants for rescue of Ecdfl/fl MEFs from cell cycle arrest induced by Cre-mediated Ecd deletion. Ecdfl/fl MEFs expressing vector (V), WT hECD or its 3S/A, 6S/A, 6S/D or 3′S/A mutants were infected with control (ctrl) or Cre adenoviruses, followed by cell counting at the indicated time points. The Cre/Ctrl cell number ratios at each time point were plotted to assess the level of rescue relative to vector-expressing MEFs. Simulation's correction was applied to control for multiple testing in the calculation of the mean ratio. The experiment is representative of three repeats.
CK2 is known to phosphorylate its substrates in clusters, with phosphorylation at one site priming the substrate for phosphorylation at additional sites (27, 28). The C-terminal region contains two potential Ser clusters: a proximal cluster of S503, S505, and S518 and a distal cluster of S572, S579, and S584 (Fig. 3C). To assess the contribution of these clusters to CK2-dependent phosphorylation of ECD, we introduced Ser-to-Ala mutations in these residues, individually as well as in combinations (Fig. 3C and D). While S→A mutations of the Ser residues in the distal cluster (S572A, S579A, and S584A; designated 3′S/A) had no appreciable impact on the level of phosphorylation in the in vitro kinase assay, similar mutations in the proximal cluster (S503A, S505A, and S518A; designated 3S/A) led to a considerable reduction in the CK2-mediated phosphorylation (Fig. 3D). Importantly, Ala mutations of all six residues (S503, S505, S518, S572, S579, and S584; designated 6S/A) nearly completely abolished the CK2-mediated in vitro phosphorylation of ECD (Fig. 3D). The autoradiography results were confirmed by subjecting the same filters to blotting with anti-p-Ser antibody (Fig. 3D). Since we did not observe a shift in the mobility of the 6S/A mutant on regular SDS-PAGE, perhaps reflecting a mechanism previously reported by Lee et al. (29), we performed gel analysis of in vitro-phosphorylated WT, 6S/A, and phosphatase-treated WT ECD after reacting these with Phos-tag, a dinuclear metal complex that acts as a phosphate-binding tag and can produce a mobility shift (30). As expected, the Phos-tag-bound WT ECD resolved as a slower-migrating band compared to its phosphatase-treated sample; notably, the 6S/A mutant exhibited a faster mobility compared to phosphorylated WT ECD (Fig. S1A). Collectively, these results show that the C-terminal Ser clusters of ECD, especially the proximal one, can be phosphorylated by CK2. The small residual phosphorylation signal observed with the 6S/A mutant of ECD may reflect an additional minor site of CK2-mediated phosphorylation.
Although these experiments confirmed and extended the concept of CK2-mediated phosphorylation of ECD in vitro, to relate this posttranslational modification to ECD function, it was important to assess whether ECD is phosphorylated in cells on the same sites and whether such phosphorylation is important for its function. Thus, we generated pMSCV-puro vector-based retroviral constructs encoding FLAG-tagged wild type or 3S/A or 6S/A mutants of ECD. These constructs were expressed in T98G cells, and cells were metabolically labeled with [32P]orthophosphate. Equal amounts of radioactive extracts (based on the counts per minute) were subjected to anti-FLAG IP, followed by autoradiography. Although the phosphorylation signal observed with the 3S/A mutant was comparable to that on the WT ECD, the level of phosphorylation on the 6S/A mutant was markedly reduced (Fig. 3E). To ascertain whether the defective phosphorylation of the cell-expressed 6S/A mutant reflects simply a lack of phosphorylation of the distal serine cluster, we compared the levels of phosphorylation of FLAG-tagged WT versus 3′S/A mutant by anti-p-Ser immunoblotting of anti-FLAG IPs of lysates of T98G cells transfected with the respective constructs. We did not observe any significant differences in the anti-p-Ser signals of WT ECD versus its 3′S/A mutant (Fig. S1B). These results establish that the two serine clusters in ECD identified in vitro as CK2 substrate sites are the major sites of phosphorylation in cells. Next, T98G cells expressing FLAG-tagged WT, 3S/A, and 6S/A ECD proteins were left untreated or treated with a CK2-specific inhibitor TBB and their anti-FLAG IPs were blotted with anti-p-Ser antibody (Fig. 3F). Notably, CK2 inhibition reduced the phosphorylation signal in cells expressing the WT ECD or its 3S/A mutant; however, cells expressing the 6S/A mutant did not exhibit any change in phosphorylation (Fig. 3F), indicating that CK2 is the primary cellular kinase responsible for the phosphorylation of ECD on two major serine clusters characterized here. However, it remains possible that the additional Ser or Thr residues of ECD are phosphorylated by other kinases, depending on the cell type or varying functional states.
Next, we assessed whether the phosphorylation of ECD on CK2-dependent serine clusters is relevant to its cell cycle regulatory function. We have previously demonstrated that introduction of Cre recombinase in Ecdfl/fl MEFs, using adenovirus Cre, causes G1 cell cycle arrest that is largely rescued by introducing human ECD (6). We used this approach to compare the extent of the rescue of cell cycle arrest induced by endogenous ECD deletion upon introducing the wild-type ECD or its phosphodefective mutants. In initial experiments, we expressed the WT human ECD or its 3S/A or 6S/A mutants in Ecdfl/fl MEFs (Fig. 3G) and then assessed the ability of the cells to progress through cell cycle without or with Cre-induced Ecd deletion. In each case, the expression of exogenous human ECD proteins and the depletion of endogenous mouse ECD were confirmed by Western blotting (Fig. 3G). As expected, the deletion of Ecd in Ecdfl/fl MEFs arrested proliferation with no recovery during the entire observation period (Fig. 3H), and ectopic WT ECD significantly rescued the cells from growth arrest (Fig. 3H; also see Fig. S2A in the supplemental material). Notably, while the 3S/A mutant behaved comparably to WT ECD in rescuing cells from growth arrest, the 6S/A mutant only exhibited a partial rescue in comparison to that seen with WT ECD in repeated experiments (P < 0.001) (Fig. 3H) (all P values are shown in Table S1 in the supplemental material). Furthermore, we examined a mutant in which serine residues 503, 505, and 518 were removed by deletion (Δ499–527) in the cell cycle rescue experiment and observed that this mutant behaved similarly to WT ECD in rescuing the proliferation block (see Fig. S2A and B in the supplemental material). Next, we generated a phosphomimetic mutant in which the six serine residues identified to be phosphorylated were mutated to aspartic acid residues (6S/D). Notably, the phosphomimetic mutant 6S/D was completely defective in cell cycle rescue experiment (Fig. 3I and J). In these experiments, we also examined the 3′S/A mutant and observed a partial rescue with this mutant (Fig. 3J). Although the complete lack of rescue seen with the 6S/D mutant of ECD was surprising, it has been reported that aspartic acid phosphomimics are unsuitable for biological readouts due to different chemical properties of the two residues (31, 32). Taken together, our results underscore the importance of ECD phosphorylation for cell cycle progression.
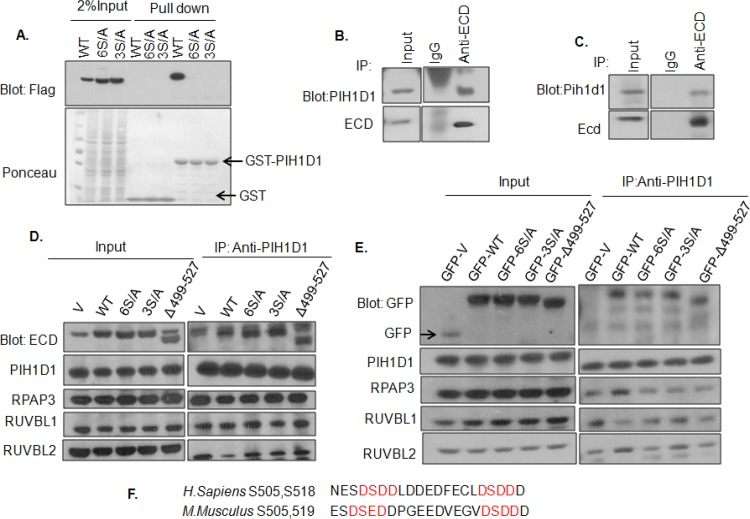
Phosphodefective ECD mutants retain their ability to interact with PIH1D1 protein, as well as with other components of the R2TP complex.
In view of the complete lack of any functional impact of mutating ECD on S503/505/518 residues, we first reexamined the previously reported dependence of ECD binding to the isolated phospho-reader domain of PIH1D1 (11). For this purpose, GST-PIH1D1 pulldown was carried out with lysates of HEK-293T cells transiently transfected to express WT ECD or its 3S/A and 6S/A mutants. Confirming previous findings (11), WT ECD, but not its 3S/A or 6S/A mutant, was pulled down with GST-PIH1D1 (Fig. 4A). These results suggested that either the R2TP association was unnecessary for ECD function in cell cycle progression or an alternate mechanism may recruit ECD to the R2TP complex. To distinguish between these possibilities, we first carried out anti-ECD IPs of U2OS and MEF cell lysates, followed by anti-PIH1D1 blotting. These analyses confirmed the interaction of endogenous ECD and PIH1D1 (Fig. 4B and C). To examine the nature of ECD/PIH1D1 interaction in cells, we carried out anti-PIH1D1 immunoprecipitations from HEK-293T cells expressing untagged (Fig. 4D) or GFP-tagged ECD (Fig. 4E; see also Fig. S3A in the supplemental material), WT ECD or its phosphorylation site mutants (defective in binding to PIH1D1 in the pulldown assay), followed by blotting for ECD (Fig. 4D) or GFP (Fig. 4E, see also Fig. S3A in the supplemental material), as well as for the four R2TP complex components (Fig. 4D and E; see also Fig. S3A in the supplemental material). Since a phosphorylated DSpDD/E motif, conserved between human and mouse ECD proteins (Fig. 4F), was previously found to promote the interaction of ECD with PIH1D1 in vitro (11), we also examined a deletion construct (Δ499–527) of ECD that lacks the DSDD/E motif in addition to the S6/A mutant lacking all CK2-phosphorylated sites. As expected, PIH1D1 IPs were able to co-IP RPAP3, RUVBL1, or RUVBL2 to a similar extent in all lanes (Fig. 4D and E). Notably, compared to the levels of endogenous ECD co-IP with PIH1D1 in vector control lanes, increased amounts of ectopically expressed ECD were coimmunoprecipitated from WT ECD-transfected cell lysates. Unexpectedly, however, the WT and mutant ECD proteins were coimmunoprecipitated with PIH1D1 to comparable levels (Fig. 4D and E; see also Fig. S3A in the supplemental material). These results demonstrate that ECD interacts with the R2TP complex in cells but that the phosphorylation-dependent interaction of ECD with PIH1D1 is dispensable for this association. In further support of this conclusion, we carried out anti-ECD and anti-PIH1D1 IPs, treated these IPs with lambda phosphatase, and then assessed the levels of coimmunoprecipitated PIH1D1. Notably, although the phosphatase treatment robustly eliminated the phosphorylation signal on ECD (anti-p-Ser blot), no reduction in PIH1D1or ECD coimmunoprecipitation was seen (see Fig. S3B and C in the supplemental material). Thus, while CK2-phosphorylated ECD can directly interact with PIH1D1, as reported previously (11), this interaction is not required for the association of ECD with the R2TP complex. Next, we examined the ability of 6S/D or 3′S/A mutants of ECD to interact with PIH1D1. For this purpose, lysates from 293T cells expressing FLAG-tagged 3′S/A or 6S/D mutants were used for an in vitro pulldown assay with GST-PIH1D1. As expected, GST-PIH1D1 was able to pull down the 3′S/A mutant but failed to pull down the 6S/D mutant of ECD (see Fig. S3D in the supplemental material), confirming that 6S/D does not mimic WT ECD for its interaction with PIH1D1.
FIG 4.
ECD phosphorylation in living cells is dispensable for its interaction with PIH1D1 and other components of the R2TP complex. (A) In vitro interaction of GST-PIH1D1 with ECD and its mutants. GST-PIH1D1 was immobilized on glutathione-Sepharose beads and incubated with lysates of HEK-293 cells expressing FLAG-tagged WT ECD or its 3S/A or 6S/A mutant. (B and C) Interaction between endogenous ECD and PIH1D1 was confirmed by immunoprecipitation of ECD from U2OS or MEFs, followed by Western blotting with anti-PIH1D1 antibody. (D and E) Lysates of HEK-293T cells transfected with untagged (D) or GFP-tagged (E) WT, 3S/A, 6S/A, or Δ499–527 ECD were subjected to immunoprecipitation with anti-PIH1D1 antibody and immunoblotted with the indicated antibodies. (F) Alignment of mouse and human ECD sequences in the region containing the DSDD/E motif. V, vector-transfected cells.
Novel phosphoindependent interaction of ECD with R2TP complex through RUVBL1.
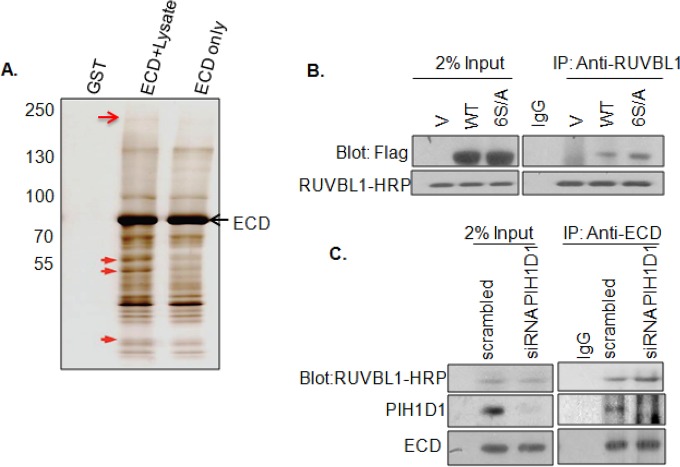
Since disabling ECD binding to PIH1D1 in the 3S/A mutant had no impact on ECD association with the R2TP complex in cells or on ECD function during cell cycle progression, we used an unbiased approach to identify potential mediators of ECD's interaction with the R2TP complex. We used an in vitro tandem affinity purification approach (23) to identify ECD interacting partners. For this purpose, full-length ECD was tagged with GST on the N terminus and with FLAG epitope on the C terminus, and the twin-tagged recombinant protein was prepared in a glutathione-Sepharose bead-bound form. Cell lysates prepared from 76NTERT cells were incubated with these beads, and proteins in the complex were eluted by cleaving the ECD-FLAG part of the GST-ECD-FLAG fusion on beads with PreScission protease. The eluted ECD-FLAG, in complex with cellular proteins, was subjected to a second round of affinity purification using anti-FLAG antibody beads, and the protein complexes were eluted with a FLAG epitope peptide. The proteins in the complex were separated by SDS-PAGE and visualized by silver staining (Fig. 5A), and bands only seen in lanes where recombinant protein was incubated with cell lysates were excised and subjected to mass spectrometry. Proteins with Mascot scores of >50 were considered potential interacting partners.
FIG 5.
ECD interacts with RUVBL1. (A) Tandem affinity purification identified RUVBL1 as an ECD-interacting protein. 76NTERT cell lysates were incubated with glutathione-Sepharose bead-bound GST-ECD-FLAG and protein complexes eluted by cleaving the ECD-FLAG portion with PreScission protease. Eluted ECD-FLAG in complex with bound proteins was further affinity purified using FLAG beads and then eluted using excess FLAG peptide. The presented gel corresponds to 5% of the final eluates visualized by silver staining. Arrows point to the gel slices that were analyzed by mass spectrometry. The top and middle arrows point to slices that identified PRPF8 and RUVBL1, respectively. (B) Interaction between endogenous RUVBL1 and ECD. Lysates of HEK-293T cells expressing FLAG-tagged WT ECD or its 6S/A mutant were subjected for anti-RUVBL1 IP, followed by anti-FLAG blotting. (C) PIH1D1 knockdown does not affect RUVBL1-ECD association. Lysates of U2OS cells transfected with PIH1D1 or scrambled control siRNA 48 h earlier were subjected to anti-ECD IP followed by blotting with antibodies against the indicated proteins. V, vector-transfected cells.
These analyses identified a known ECD binding partner PRPF8 (7) and in addition revealed several new binding partners. Interaction of PRPF8 with ECD was confirmed by immunoprecipitation (see Fig. S3E in the supplemental material). Among the new partners, RUVBL1 was one of the top candidate proteins with a Mascot score of 168. To validate the purification results, we expressed the FLAG-tagged WT ECD or its 6S/A mutant in 293T cells and performed coimmunoprecipitation experiments with an anti-RUVBL1 antibody. Both the WT and the 6S/A ECD proteins were coimmunoprecipitated with RUVBL1, suggesting that ECD interacts with RUVBL1 and that this interaction is independent of ECD phosphorylation (Fig. 5B). To further establish that ECD-RUVBL1 interaction is independent of PIH1D1, we knocked down the endogenous PIH1D1 with siRNA and then performed a co-IP experiment using an anti-ECD antibody. Notably, we observed equal co-IP of RUVBL1 in both control and PIH1D1 knockdown cells, confirming that ECD interaction with RUVBL1 is PIH1D1 independent (Fig. 5C). Taken together, these results demonstrate that ECD associates with the R2TP complex though a novel interaction with RUVBL1, independent of ECD's interaction with PIH1D1.
Interaction with RUVBL1 is important for the role of ECD in cell cycle progression.
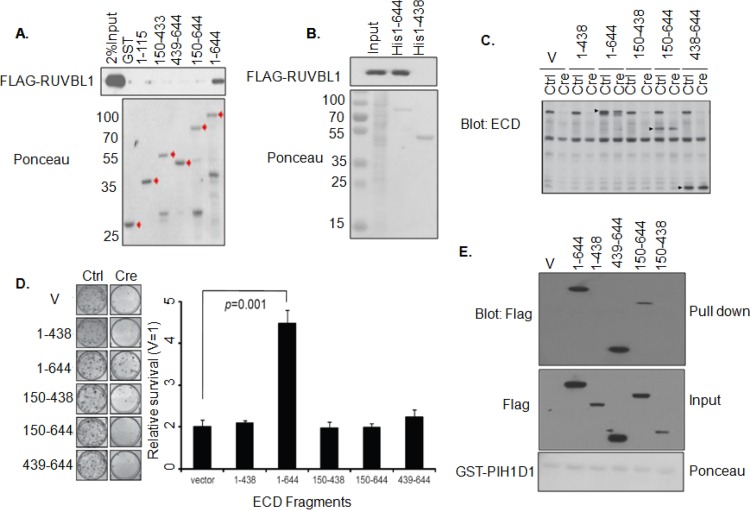
Germ line deletion of Ecd or Ruvbl1 is embryonic lethal (6, 18), and silencing of either protein in cells leads to cell cycle arrest (6, 17, 18), suggesting that interaction of ECD with RUVBL1 may play a role in the cell cycle regulation function of ECD. To test this hypothesis, we first expressed GFP-tagged ECD or its several C-terminal deletion mutants in HEK-293T cells and performed co-IP experiments using an anti-RUVBL1 antibody. Notably, only full-length ECD protein (amino acids [aa] 1 to 644) coimmunoprecipitated with RUVBL1, whereas none of the C-terminal deletions of ECD was able to coimmunoprecipitate with RUVBL1 (see Fig. S4A and B in the supplemental material). To validate these results, we constructed several GST-tagged and His-tagged ECD deletion fragments based on the predicted secondary structure (by Garnier Robson predictions and PONDR VL-XT secondary structure prediction) and then examined the direct interaction of these ECD mutant proteins with FLAG-RUVBL1 or endogenous RUVBL1 using pulldown with glutathione-Sepharose or nickel beads. As shown in (Fig. 6A and B; see also Fig. S4C in the supplemental material), only the full-length ECD interacts with RUVBL1, whereas all C-terminal or N-terminal deletions rendered ECD defective in binding to RUVBL1. Next, we compared various FLAG-tagged deletion fragments of ECD (residues 150 to 438, 438 to 644, 1 to 438, and 150 to 644) (see Fig. S4D in the supplemental material) with wild-type ECD (residues 1 to 644) for their abilities to rescue the growth arrest of Ecdfl/fl MEFs upon adeno-Cre-mediated endogenous Ecd deletion by analyzing cell proliferation by cell counting or colony formation (Fig. 6D). In each case, the expression of exogenous human ECD proteins and loss of expression of endogenous mouse Ecd in Cre-expressing cells was confirmed using Western blotting (Fig. 6C; see also Fig. S4D in the supplemental material). As expected, deletion of Ecd in Ecdfl/fl MEFs arrested cell proliferation, and ectopic WT human ECD significantly rescued the cells from growth arrest (Fig. 6D). However, none of the deletion mutants was able to rescue the cell proliferation block imposed by endogenous ECD depletion. These results demonstrate that only the full-length ECD, which interacts with RUVBL1, supports cell cycle progression. The lack of rescue with ECD deletion fragments was not due to lack of their expression (Fig. 6C; see also Fig. S4D in the supplemental material). Notably, two deletion fragments (residues 438 to 644 and 150 to 644) that failed to rescue cell cycle arrest still retained their ability to interact with PIH1D1 (Fig. 6E), further underscoring the conclusion that interaction of ECD with PIH1D1 is dispensable, while its interaction with RUVBL1 is indispensable for a role in cell cycle progression. PIH1D1 is known to directly interact with other components of the R2TP complex, such as RUVBL1 (33). Reprobing of the same membrane with antibodies against RUVBL1 and RUVBL2 showed the expected interaction of PIH1D1 with RUVBL1 or RUVBL2 (see Fig. S4E in the supplemental material).
FIG 6.
Interaction with RUVBL1 is important for ECD function in cell cycle progression. (A and B) Interaction between FLAG-RUVBL1 and ECD. GST-tagged or His-tagged full-length ECD or its truncated mutants immobilized on glutathione-Sepharose or nickel beads, respectively, were incubated with lysates of HEK-293T cells expressing FLAG-tagged RUVBL1. (C) Western blotting to show the expression of WT human ECD or its deletion mutants in Ecdfl/fl MEFs with control or Cre adenovirus infection (arrowheads point to the human ECD or mutants). Also note that anti-ECD antibody blot does not detect the C-terminally deleted ECD 1-438 mutant. (D) Colony formation assay. Ecdfl/fl MEFs expressing full-length WT ECD (aa 1 to 644) or its truncations were infected with ctrl or Cre adenoviruses, colonies were stained with crystal violet after 10 days, and the solubilized dye absorbance was read at 590 nm. A histogram shows the relative rescue efficiency of each construct compared to vector control cells. Error bars represent the means ± the standard deviations of three independent experiments. A statistical comparison used a Student two-tailed t test. (E) Interaction of FLAG-tagged ECD or its deletion fragments with PIH1D1. Lysates of HEK-293T cells expressing the indicated FLAG-tagged ECD fragments were used for GST-PIH1D1 pulldown, followed by anti-FLAG blotting. V, vector-transfected cells.
Our previous studies showed that ECD interacts with RB, a function important for the ECD role in cell cycle progression (6). Notably, in addition to the expected interaction of full-length ECD with RB, one mutant (aa 150 to 644) that is defective in rescuing cell cycle arrest (Fig. 6D) was earlier shown to retain its ability to interact with RB (6), suggesting that interaction with RUVBL1 is required for ECD to promote cell cycle progression while interaction with RB in the absence of interaction with RUVBL1 is insufficient for this function. In further support of this conclusion cell cycle function competent (3S/A) and deficient mutant (6S/A) of ECD show comparable interaction with RB (see Fig. S4F in the supplemental material).
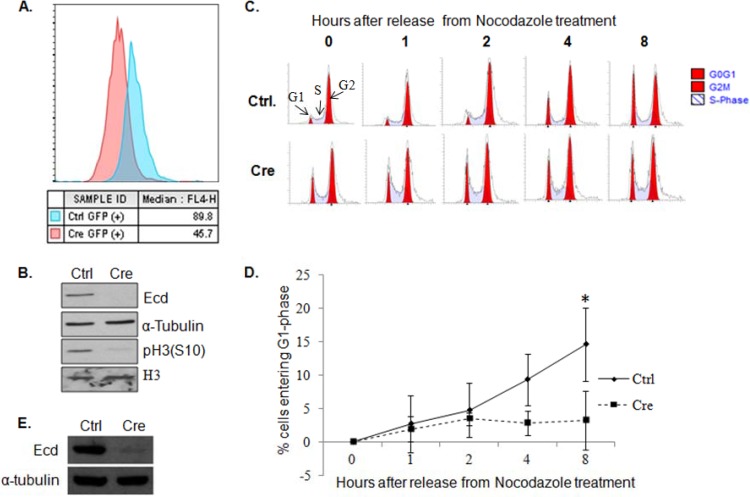
Ecd deletion leads to reduced mitotic index and delayed mitotic progression.
We have previously reported that the proliferation arrest upon Ecd deletion is not associated with any increase in apoptosis (6). To examine the effect of Ecd deletion on mitosis, we used adeno-Cre to delete Ecd in Ecdfl/fl MEFs and measured the MFI of phospho-histone H3 (S10) as an indicator of the proportion of cells in mitosis using flow cytometry (34). Ecd-deleted cells showed a marked decrease in the MFI (45.7) of pH 3 (S10) compared to control cells (89.8) (Fig. 7A), indicating that Ecd deletion cells are cell cycle arrested prior to entering mitosis. Low levels of pH 3 (S10) were further confirmed by Western blotting (Fig. 7B). Next, we assessed the G2/M to G1 progression of MEFs arrested in the S phase by nocodazole treatment (Fig. 7C). Flow cytometry analysis revealed a significant impairment in G2/M-to-G1-phase transition upon Ecd deletion compared to control, in addition to a higher percentage of Ecd-deleted MEFs in the G1 phase (Fig. 7C to E). Taken together, these results demonstrate a critical role of ECD in both G1-to-S and G2/M-to-G1 transitions. These results are consistent with the known function of CK2 and RUVBL1 in cell cycle regulation (35–37).
FIG 7.
Ecd deletion leads to a mitotic block. (A) Ecdflox/flox MEFs infected with control (blue) or Cre adenoviruses (red) were fixed in 70% ethanol, stained with anti-pH3(S10), and analyzed by flow cytometry. The median fluorescence intensity (MFI), representing the peak channel number on the x axis, is shown. (B) Lysates of control (Ctrl) or Ecd-deleted (Cre) Ecdflox/flox MEFs were blotted with the indicated antibodies. (C) Control (Ctrl) or Ecd deletion (Cre) MEFs were treated with 100 ng of nocodazole/ml for 20 h, and cells were switched to nocodazole-free medium to initiate cell cycle progression for the indicated time points. The cells were stained with propidium iodide and analyzed by FACS for cell cycle analysis. (D) A graph shows the percentage of cells entering the G1 phase after release from nocodazole treatment at various time points. Data points are means ± the standard deviations of results from three independent experiments (*, P ≤ 0.05, as determined by two-tailed Student t test). (E) The deletion of Ecd in the experiment shown in panel C was confirmed by Western blotting.
DISCUSSION
Precise regulation of the entry into, progression through, and exit from the cell cycle is fundamental to developmental programs and maintenance of adult tissues in multicellular organisms. Notably, components of the cell cycle machinery and the pathways that regulate their functions are commonly altered in cancer and other diseases (1). Thus, elucidating how the cell cycle machinery is controlled is an important area of research in cell and cancer biology.
We have previously shown that ECD, the mammalian orthologue of Drosophila ecdysoneless gene, is required for embryonic development and progression of mammalian cells through the G1-S phase of cell cycle progression (6). Here, we identify a novel mechanism by which ECD functions as an essential element of mammalian cell cycle progression. Using multiple complementary approaches, we demonstrate a novel interaction of ECD with the R2TP chaperone complex, mediated by the RUVBL1 component of R2TP, which we establish is required for ECD to promote cell cycle progression. We also identify a role for the CK2-dependent phosphorylation of ECD in cell cycle progression. In contrast to predictions from a previous study (11), this role is independent of the ECD interaction with PIH1D1, the phospho-reader component of the R2TP complex.
Our findings establish that the phosphorylation of ECD positively regulates its function in promoting the cell cycle progression. Bioinformatics analysis, followed by mass spectroscopy-based phosphoproteomics, identified a number of sites that could be phosphorylated by cellular kinases, but we focused on two clusters of potential CK2 phosphorylated serine residues since a recent study (11) showed that CK2-mediated phosphorylation of two such serine residues in the context of a peptide created a binding site for the phospho-reader subunit of the R2TP complex. CK2-dependent phosphorylation of site-directed mutants of ECD in vitro and in cultured cells identified six serine residues in two spatially separated clusters to be the major CK2 phosphorylation sites on ECD. Notably, however, ECD phosphorylation does not change during cell cycle progression. This is not entirely surprising since our in vitro analyses, as well as phosphorylation studies in cells in the presence of a CK2 inhibitor (Fig. 3D and F), establish that CK2 is the predominant kinase that phosphorylates ECD; CK2 is considered to be constitutively active and ubiquitous serine/threonine protein kinase (38). Despite its constitutive activity, however, numerous studies point to a role for CK2 in cell proliferation and survival (39). Yet, the molecular pathways that mediate the function of CK2 in cell proliferation are largely unknown. We suggest that phosphorylation of ECD by CK2 provides one mechanism for CK2's role in cell proliferation. Although the overall levels and the subcellular localization of ECD remain invariant during cell cycle progression (Fig. 1 and 2), it remains possible that ECD phosphorylation at specific sites may vary during cell cycle progression. As phospho-specific antibodies against specific serine residues on ECD become available, it should be feasible to test this notion further.
Our findings that ECD is indeed a CK2 substrate in vitro (Fig. 3) suggested that CK2-dependent phosphorylation and subsequent interaction of ECD with the R2TP complex could provide a potential mechanism by which ECD could promote the cell cycle transit. Our co-IP studies in cell cultures demonstrate that ECD in fact is in a complex that includes the four core subunits of the R2TP complex (Fig. 4D and E). Remarkably, however, multiple mutant ECD proteins, rendered incapable of directly interacting with PIH1D1, including mutations of critical serine residues in the 3S/A mutant or the 6S/A mutant or deletion of the region incorporating the major CK2 phosphorylation sites and the acidic motif DSDD that facilitates PIH1D1 interaction (11), fully retained the ability to associate with the R2TP complex. Furthermore, the ECD-R2TP association was retained in PIH1D1-depleted cells (Fig. 5C). Thus, our results support a PIH1D1-independent mechanism of ECD association with the R2TP complex. Importantly, S→A mutation of ECD residues that impart PIH1D1 binding (3S/A and Δ499–527) had no impact on its ability to function in cell cycle progression. However, 6S/A and 6S/D were defective in a cell cycle rescue experiment, underscoring the importance of ECD phosphorylation for its function that encompasses amino acids beyond PIH1D1 interaction. Thus, the role of phosphorylation in regulating ECD function during cell cycle appears to be independent of mediating an interaction with PIH1D1. It remains possible, however, that phosphorylation-dependent interaction of ECD with PIH1D1, and consequently with the R2TP complex, is required for other functions of ECD aside from its role in promoting cell cycle progression (6). We have shown that ECD overexpression in cells leads to p53 stabilization and increased p53-dependent target gene expression and to the induction of a senescence phenotype in primary fibroblasts (5). ECD was also found to interact with thioredoxin-interacting protein (TXNIP), which was shown to promote p53 stabilization (40). TXNIP has a number of other functions, including the regulation of glucose uptake, oxidative stress, and endoplasmic reticulum stress-induced apoptosis (41, 42), Thus, ECD phosphorylation and interaction with PIH1D1 may play a role in regulating these functions. The availability of Ecdfl/fl MEFs in which ECD can be conditionally deleted, together with the phosphorylation-defective mutants that we have characterized here, should allow these notions to be tested in the future.
In view of a novel, PIH1D1-independent mechanism of ECD association with the R2TP complex, we sought to answer two key mechanistic questions: first, what are the determinants of ECD-R2TP association, and second, whether this unique mode of interaction is functionally relevant in the context of cell cycle progression role of ECD. Unbiased proteomics analysis of cellular proteins that interacted with a recombinant full-length ECD protein, followed by biochemical analyses in cells, demonstrated that ECD interacts with another component of the R2TP complex, RUVBL1 (Fig. 5 and 6). Structure-function studies of ECD using deletion fragments demonstrated a strong correlation between the cell cycle progression function of ECD and its ability to interact with RUVBL1, with only the full-length ECD competent at both functions (Fig. 6A and B; see also Fig. S4B and C in the supplemental material). Interestingly, the Δ499–527 mutant which interacts with RUVBL1, but not PIH1D1, was able to rescue the cell cycle arrest caused by Ecd deletion (see Fig. S2A in the supplemental material). Thus, our studies identify a novel interaction of ECD with RUVBL1 and suggest that this mode of interaction with the R2TP complex is a key to the regulation of cell cycle progression by ECD. The delineation of sequences in ECD and RUVBL1 that mediate their interaction should help directly test whether selective abrogation of this interaction is functionally critical in cell cycle progression, as well as to assess the potential role of ECD in other roles of RUVBL1 within the R2TP complex. Interestingly, mouse ECD and RUVBL1 knockouts are phenotypically similar, since both are embryonic lethal at the blastocyst stage (6, 19). RUVBL1 is essential for cellular proliferation as seen in knockout cells or upon knockdown of RUVBL1 expression (18). A recent study demonstrated that RUVBL1 functions as a critical factor for p300 recruitment to OCT4 target genes (18). It is of interest that ECD also interacts with p300 and promotes its transcriptional coactivator function (8). Thus, ECD may function in close coordination with RUVBL1.
Given the evidence we present that ECD can physically interact with two distinct components of the R2TP complex, it is conceivable that certain ECD functions require both modes of interaction. Recent studies have shown that, aside from the R2TP complex, RUVBL1/2 are also parts of other functionally relevant complexes, such as chromatin remodeling complexes TIP60, SWR/SRCAP, and INO80 and the Fanconi anemia core complex that controls DNA interstrand cross-link repair and function, and regulate telomerase biogenesis and mitosis (19, 43–45). Given the PIH1D1-independent interaction of ECD with RUVBL1, the potential roles of ECD via these alternative complexes will be of great future interest.
An essential role of ECD in cell cycle progression was established by our previous observation that ECD is essential for embryogenesis and its conditional deletion in MEFs leads to a G1-S cell cycle arrest, together with an inability to initiate an E2F-dependent transcriptional program essential for cell cycle progression (6). Notably, we demonstrated that ECD competes with E2F for binding to the pocket domain of RB and that the cell cycle progression defect in Ecd-null MEFs could be overcome by removing the RB-mediated suppression of E2F using a pocket-binding oncogene HPV16 E7. Since a key mechanism by which the R2TP complex regulates biochemical processes is by facilitating protein complex remodeling, we speculate that the interaction of ECD with the R2TP complex through RUVBL1 may facilitate the ECD-RB complex formation and helps dissociate RB from E2Fs, thereby derepressing the E2F-mediated transcription and promoting cell cycle progression. Consistent with this speculative model, our previous studies showed that binding to RB was not sufficient for the cell cycle progression function of ECD, since we identified one ECD mutant that was able to interact with RB but was defective in cell cycle rescue.
Our previous studies demonstrated that ECD is overexpressed in breast and pancreatic cancer patient tissues and that ECD overexpression correlates with poor prognosis and poor survival in breast cancer patients (9, 10). It is noteworthy that several components of the R2TP/prefoldin complex, including PIH1D1, RUVBL1, and RUVBL2, are also overexpressed in various cancers and are predicted to play important roles in oncogenesis (46, 47). A comprehensive meta-analysis of The Cancer Genome Atlas (TCGA) data sets (46) revealed that expression of many RUVBL complex genes was significantly higher in breast and colorectal carcinomas compared to their normal tissue controls. These investigations suggested a correlation between RUVBL complex component overexpression and increased mTORC1 signaling and metabolic processes necessary for tumor cell growth (46). Another study demonstrated that PIH1D1 is overexpressed in various breast cancer cell lines, where it plays a major role in rRNA transcription (48). Our recent studies showed a co-oncogenic role of ECD with Ras when introduced into immortal human mammary epithelial cells (49), further suggesting the potential collaborative role of ECD and the R2TP or other RUVBL-containing complexes in cell cycle regulation and oncogenesis.
A positive role of ECD in pre-mRNA splicing was reported recently based on rescue of splicing defects in the prothoracic glands of Ecd-deficient flies by human ECD and interaction of ECD with a complex containing the spliceosome component PRP8 (7, 50). Our affinity purification/mass spectrometry analyses confirmed the interaction of ECD with PRPF8. The R2TP complex regulates mRNA and ribosome biogenesis by facilitating the assembly of small nucleolar ribonucleoproteins (snoRNPs), which are known to be involved in spliceosome modification (51, 52). Upregulation of R2TP and snoRNP components is thought to promote ribosome synthesis in cancer cells (47). Whether overexpressed ECD in tumors may function in concert with R2TP and other RUVBL1-containing complexes to promote oncogenesis requires further investigation. Taken together, the findings presented here demonstrate that CK2-mediated phosphorylation and interaction with RUVBL1 are essential for ECD's ability to regulate cell cycle progression.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Band laboratories for their thoughtful discussions and suggestions throughout this work, James H. Prestegard (University of Georgia) for kindly providing the ST6GAL1-pXLG-NtermTCMhisStrep-DEST construct, and the UNMC Protein Structure Core Facility for help with protein purification.
Funding Statement
Work in our laboratories is supported by National Institutes of Health (NIH) grants CA96844 and CA144027 to V.B. and CA87986 and CA99163 to H.B., by Department of Defense grants W81XWH-07-1-0351, W81XWH-11-1-0171, and W81XWH-14-1-0567 to V.B., and by a Fred & Pamela Buffett Cancer Center Support Grant (P30CA036727). S.M. was supported by Susan G. Komen postdoctoral fellowship grant KG111248. A.B. was supported by DOD BCRP predoctoral fellowship grant W8X1XWH-11-0020.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00594-15.
REFERENCES
- 1.Hanahan D, Weinberg RA. 2000. The hallmarks of cancer. Cell 100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Garen A, Kauvar L, Lepesant JA. 1977. Roles of ecdysone in Drosophila development. Proc Natl Acad Sci U S A 74:5099–5103. doi: 10.1073/pnas.74.11.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaziova I, Bonnette PC, Henrich VC, Jindra M. 2004. Cell-autonomous roles of the ecdysoneless gene in Drosophila development and oogenesis. Development 131:2715–2725. doi: 10.1242/dev.01143. [DOI] [PubMed] [Google Scholar]
- 4.Sato T, Jigami Y, Suzuki T, Uemura H. 1999. A human gene, hSGT1, can substitute for GCR2, which encodes a general regulatory factor of glycolytic gene expression in Saccharomyces cerevisiae. Mol Gen Genet 260:535–540. doi: 10.1007/s004380050926. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Chen J, Gurumurthy CB, Kim J, Bhat I, Gao Q, Dimri G, Lee SW, Band H, Band V. 2006. The human orthologue of Drosophila Ecdysoneless protein interacts with p53 and regulates its function. Cancer Res 66:7167–7175. doi: 10.1158/0008-5472.CAN-06-0722. [DOI] [PubMed] [Google Scholar]
- 6.Kim JH, Gurumurthy CB, Naramura M, Zhang Y, Dudley AT, Doglio L, Band H, Band V. 2009. Role of mammalian Ecdysoneless in cell cycle regulation. J Biol Chem 284:26402–26410. doi: 10.1074/jbc.M109.030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claudius AK, Romani P, Lamkemeyer T, Jindra M, Uhlirova M. 2014. Unexpected role of the steroid-deficiency protein Ecdysoneless in pre-mRNA splicing. PLoS Genet 10:e1004287. doi: 10.1371/journal.pgen.1004287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JH, Gurumurthy CB, Band H, Band V. 2010. Biochemical characterization of human Ecdysoneless reveals a role in transcriptional regulation. Biol Chem 391:9–19. doi: 10.1515/BC.2010.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X, Mirza S, Alshareeda A, Zhang Y, Gurumurthy CB, Bele A, Kim JH, Mohibi S, Goswami M, Lele SM, West W, Qiu F, Ellis IO, Rakha EA, Green AR, Band H, Band V. 2012. Overexpression of a novel cell cycle regulator Ecdysoneless in breast cancer: a marker of poor prognosis in HER2/neu-overexpressing breast cancer patients. Breast Cancer Res Treat 134:171–180. doi: 10.1007/s10549-011-1946-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dey P, Rachagani S, Chakraborty S, Singh PK, Zhao X, Gurumurthy CB, Anderson JM, Lele S, Hollingsworth MA, Band V, Batra SK. 2012. Overexpression of Ecdysoneless in pancreatic cancer and its role in oncogenesis by regulating glycolysis. Clin Cancer Res 18:6188–6198. doi: 10.1158/1078-0432.CCR-12-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horejsi Z, Stach L, Flower TG, Joshi D, Flynn H, Skehel JM, O'Reilly NJ, Ogrodowicz RW, Smerdon SJ, Boulton SJ. 2014. Phosphorylation-dependent PIH1D1 interactions define substrate specificity of the R2TP cochaperone complex. Cell Rep 7:19–26. doi: 10.1016/j.celrep.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakihara Y, Houry WA. 2012. The R2TP complex: discovery and functions. Biochim Biophys Acta 1823:101–107. doi: 10.1016/j.bbamcr.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Boulon S, Bertrand E, Pradet-Balade B. 2012. HSP90 and the R2TP co-chaperone complex: building multi-protein machineries essential for cell growth and gene expression. RNA Biol 9:148–154. doi: 10.4161/rna.18494. [DOI] [PubMed] [Google Scholar]
- 14.Horejsi Z, Takai H, Adelman CA, Collis SJ, Flynn H, Maslen S, Skehel JM, de Lange T, Boulton SJ. 2010. CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol Cell 39:839–850. doi: 10.1016/j.molcel.2010.08.037. [DOI] [PubMed] [Google Scholar]
- 15.Zhao R, Kakihara Y, Gribun A, Huen J, Yang G, Khanna M, Costanzo M, Brost RL, Boone C, Hughes TR, Yip CM, Houry WA. 2008. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J Cell Biol 180:563–578. doi: 10.1083/jcb.200709061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matias PM, Baek SH, Bandeiras TM, Dutta A, Houry WA, Llorca O, Rosenbaum J. 2015. The AAA+ proteins Pontin and Reptin enter adult age: from understanding their basic biology to the identification of selective inhibitors. Front Mol Biosci 2:17. doi: 10.3389/fmolb.2015.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breig O, Bras S, Martinez Soria N, Osman D, Heidenreich O, Haenlin M, Waltzer L. 2014. Pontin is a critical regulator for AML1-ETO-induced leukemia. Leukemia 28:1271–1279. doi: 10.1038/leu.2013.376. [DOI] [PubMed] [Google Scholar]
- 18.Boo K, Bhin J, Jeon Y, Kim J, Shin HJ, Park JE, Kim K, Kim CR, Jang H, Kim IH, Kim VN, Hwang D, Lee H, Baek SH. 2015. Pontin functions as an essential coactivator for Oct4-dependent lincRNA expression in mouse embryonic stem cells. Nat Commun 6:6810. doi: 10.1038/ncomms7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajendra E, Garaycoechea JI, Patel KJ, Passmore LA. 2014. Abundance of the Fanconi anaemia core complex is regulated by the RuvBL1 and RuvBL2 AAA+ ATPases. Nucleic Acids Res 42:13736–13748. doi: 10.1093/nar/gku1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Band V, Sager R. 1989. Distinctive traits of normal and tumor-derived human mammary epithelial cells expressed in a medium that supports long-term growth of both cell types. Proc Natl Acad Sci U S A 86:1249–1253. doi: 10.1073/pnas.86.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barb AW, Meng L, Gao Z, Johnson RW, Moremen KW, Prestegard JH. 2012. NMR characterization of immunoglobulin G Fc glycan motion on enzymatic sialylation. Biochemistry 51:4618–4626. doi: 10.1021/bi300319q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohibi S, Gurumurthy CB, Nag A, Wang J, Mirza S, Mian Y, Quinn M, Katafiasz B, Eudy J, Pandey S, Guda C, Naramura M, Band H, Band V. 2012. Mammalian alteration/deficiency in activation 3 (Ada3) is essential for embryonic development and cell cycle progression. J Biol Chem 287:29442–29456. doi: 10.1074/jbc.M112.378901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kataria R, Xu X, Fusetti F, Keizer-Gunnink I, Jin T, van Haastert PJ, Kortholt A. 2013. Dictyostelium Ric8 is a nonreceptor guanine exchange factor for heterotrimeric G proteins and is important for development and chemotaxis. Proc Natl Acad Sci U S A 110:6424–6429. doi: 10.1073/pnas.1301851110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisher D, Krasinska L, Coudreuse D, Novak B. 2012. Phosphorylation network dynamics in the control of cell cycle transitions. J Cell Sci 125:4703–4711. doi: 10.1242/jcs.106351. [DOI] [PubMed] [Google Scholar]
- 25.Ren S, Rollins BJ. 2004. Cyclin C/cdk3 promotes Rb-dependent G0 exit. Cell 117:239–251. doi: 10.1016/S0092-8674(04)00300-9. [DOI] [PubMed] [Google Scholar]
- 26.Su Q, Wang S, Gao HQ, Kazemi S, Harding HP, Ron D, Koromilas AE. 2008. Modulation of the eukaryotic initiation factor 2 alpha-subunit kinase PERK by tyrosine phosphorylation. J Biol Chem 283:469–475. doi: 10.1074/jbc.M704612200. [DOI] [PubMed] [Google Scholar]
- 27.St-Denis N, Gabriel M, Turowec JP, Gloor GB, Li SS, Gingras AC, Litchfield DW. 2015. Systematic investigation of hierarchical phosphorylation by protein kinase CK2. J Proteomics 118:49–62. doi: 10.1016/j.jprot.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 28.Kappes F, Damoc C, Knippers R, Przybylski M, Pinna LA, Gruss C. 2004. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol Cell Biol 24:6011–6020. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee CR, Park YH, Kim M, Kim YR, Park S, Peterkofsky A, Seok YJ. 2013. Phosphorylation-dependent mobility shift of proteins on SDS-PAGE is due to decreased binding of SDS. Bull Korean Chem Soc 88:473–485. [Google Scholar]
- 30.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. 2006. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5:749–757. [DOI] [PubMed] [Google Scholar]
- 31.de Chiara C, Menon RP, Strom M, Gibson TJ, Pastore A. 2009. Phosphorylation of S776 and 14-3-3 binding modulate ataxin-1 interaction with splicing factors. PLoS One 4:e8372. doi: 10.1371/journal.pone.0008372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menon RP, Soong D, de Chiara C, Holt MR, Anilkumar N, Pastore A. 2012. The importance of serine 776 in Ataxin-1 partner selection: a FRET analysis. Sci Rep 2:919. doi: 10.1038/srep00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jady BE, Rothe B, Pescia C, Robert MC, Kiss T, Bardoni B, Krol A, Branlant C, Allmang C, Bertrand E, Charpentier B. 2008. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs though conserved machinery. J Cell Biol 180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansen KM, Johansen J. 2006. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res 14:393–404. doi: 10.1007/s10577-006-1063-4. [DOI] [PubMed] [Google Scholar]
- 35.Pepperkok R, Lorenz P, Ansorge W, Pyerin W. 1994. Casein kinase II is required for transition of G0/G1, early G1, and G1/S phases of the cell cycle. J Biol Chem 269:6986–6991. [PubMed] [Google Scholar]
- 36.Homma MK, Homma Y. 2005. Regulatory role of CK2 during the progression of cell cycle. Mol Cell Biochem 274:47–52. doi: 10.1007/s11010-005-3111-3. [DOI] [PubMed] [Google Scholar]
- 37.Gentili C, Castor D, Kaden S, Lauterbach D, Gysi M, Steigemann P, Gerlich DW, Jiricny J, Ferrari S. 2015. Chromosome missegregation associated with RUVBL1 deficiency. PLoS One 10:e0133576. doi: 10.1371/journal.pone.0133576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meggio F, Pinna LA. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J 17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 39.Ahmed K, Gerber DA, Cochet C. 2002. Joining the cell survival squad: an emerging role for protein kinase CK2. Trends Cell Biol 12:226–230. doi: 10.1016/S0962-8924(02)02279-1. [DOI] [PubMed] [Google Scholar]
- 40.Suh HW, Yun S, Song H, Jung H, Park YJ, Kim TD, Yoon SR, Choi I. 2013. TXNIP interacts with hEcd to increase p53 stability and activity. Biochem Biophys Res Commun 438:264–269. doi: 10.1016/j.bbrc.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 41.Shalev A. 2014. Minireview: Thioredoxin-interacting protein: regulation and function in the pancreatic beta-cell. Mol Endocrinol 28:1211–1220. doi: 10.1210/me.2014-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elgort MG, O'Shea JM, Jiang Y, Ayer DE. 2010. Transcriptional and translational downregulation of thioredoxin interacting protein is required for metabolic reprogramming during G1. Genes Cancer 1:893–907. doi: 10.1177/1947601910389604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenbaum J, Baek SH, Dutta A, Houry WA, Huber O, Hupp TR, Matias PM. 2013. The emergence of the conserved AAA+ ATPases Pontin and Reptin on the signaling landscape. Sci Signal 6:mr1. doi: 10.1126/scisignal.2003906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. 2008. Identification of ATPases Pontin and Reptin as telomerase components essential for holoenzyme assembly. Cell 132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magalska A, Schellhaus AK, Moreno-Andres D, Zanini F, Schooley A, Sachdev R, Schwarz H, Madlung J, Antonin W. 2014. RuvB-like ATPases function in chromatin decondensation at the end of mitosis. Dev Cell 31:305–318. doi: 10.1016/j.devcel.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 46.Kim SG, Hoffman GR, Poulogiannis G, Buel GR, Jang YJ, Lee KW, Kim BY, Erikson RL, Cantley LC, Choo AY, Blenis J. 2013. Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol Cell 49:172–185. doi: 10.1016/j.molcel.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kakihara Y, Saeki M. 2014. The R2TP chaperone complex: its involvement in snoRNP assembly and tumorigenesis. Biomol Concepts 5:513–520. doi: 10.1515/bmc-2014-0028. [DOI] [PubMed] [Google Scholar]
- 48.Kamano Y, Saeki M, Egusa H, Kakihara Y, Houry WA, Yatani H, Kamisaki Y. 2013. PIH1D1 interacts with mTOR complex 1 and enhances ribosome RNA transcription. FEBS Lett 587:3303–3308. doi: 10.1016/j.febslet.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 49.Bele A, Mirza S, Zhang Y, Mir R, Lin S, Kim JH, Gurumurthy CB, West W, Qiu F, Band H, Band V. 2015. The cell cycle regulator Ecdysoneless cooperates with H-Ras to promote oncogenic transformation of human mammary epithelial cells. Cell Cycle 14:990–1000. doi: 10.1080/15384101.2015.1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grainger RJ, Beggs JD. 2005. Prp8 protein: at the heart of the spliceosome. RNA 11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bizarro J, Charron C, Boulon S, Westman B, Pradet-Balade B, Vandermoere F, Chagot ME, Hallais M, Ahmad Y, Leonhardt H, Lamond A, Manival X, Branlant C, Charpentier B, Verheggen C, Bertrand E. 2014. Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control. J Cell Biol 207:463–480. doi: 10.1083/jcb.201404160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bizarro J, Dodre M, Huttin A, Charpentier B, Schlotter F, Branlant C, Verheggen C, Massenet S, Bertrand E. 2015. NUFIP and the HSP90/R2TP chaperone bind the SMN complex and facilitate assembly of U4-specific proteins. Nucleic Acids Res 43:8973–8989. doi: 10.1093/nar/gkv809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.