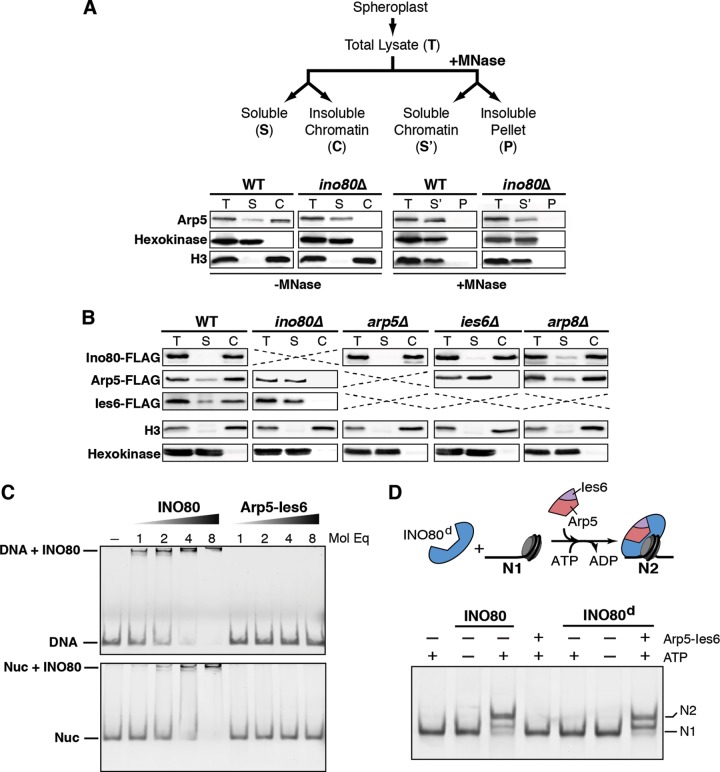
FIG 3.
Arp5-Ies6 subcomplex associates with chromatin in an INO80-dependent manner and modulates nucleosome sliding. (A) The illustration at the top shows the experimental design for in vivo chromatin fractionation with micrococcal nuclease (MNase). Below is the chromatin fractionation of wild-type (WT) and ino80Δ cells. Arp5 was detected by anti-Arp5 antibody. Hexokinase and histone H3 are indicative of cytoplasmic and chromatin fractions, respectively. MNase treatment solubilizes H3 chromatin fraction. (B) In vivo chromatin fractionation of WT and indicated deletion strains expressing Ino80-FLAG, Arp5-FLAG, or Ies6-FLAG. Arp5 and Ies6 are predominantly present in the chromatin fraction in WT cells and the soluble fraction in ino80Δ cells. Ies6-FLAG protein is undetectable in arp5Δ strains, as shown in Fig. 1D. A strain with both Ies6-FLAG and arp8Δ is nonviable. (C) Native PAGE gels of DNA (left panel) and nucleosome containing 60 bp extranucleosomal DNA (Nuc, right panel). A 2 nM concentration of DNA or nucleosome was incubated with the indicated molar equivalents (Mol Eq) of INO80 complex or Arp5-Ies6 subcomplex. (D) The illustration at the top depicts the experimental setup, wherein INO80 complex deficient of the Arp5-Ies6 subcomplex (INO80d) is preincubated with end-positioned (N1) nucleosomal substrate. Upon addition of the Arp5-Ies6 subcomplex and ATP, INO80d activity is stimulated, and the nucleosome is moved to the center position (N2).