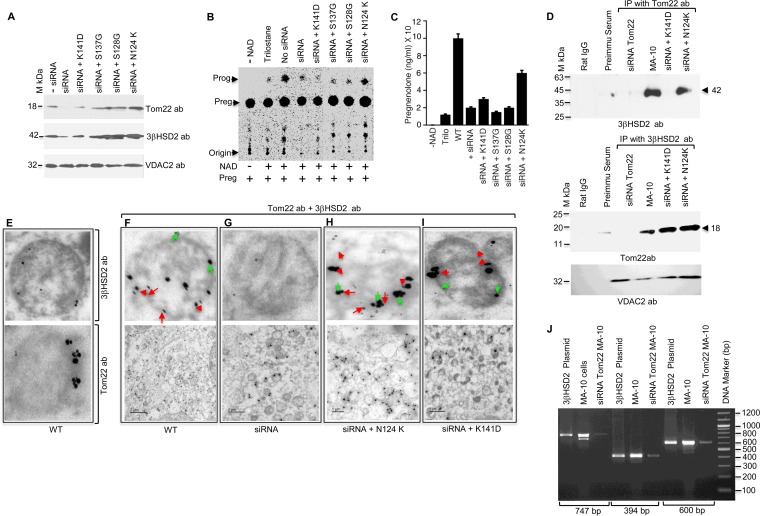
FIG 6.
Role of the C terminus of Tom22 in regulating 3βHSD2 metabolic activity. (A) The indicated Tom22 point mutants were expressed in MA-10 cells after knocking down Tom22 with 60 pmol of siRNA. Western blot analysis was performed using Tom22, 3βHSD2, and VDAC2 antibodies, confirming the equal expression of each. Expression remained unchanged with negative (−) siRNA. (B) Pregnenolone-to-progesterone conversion by ΔTom22 MA-10 cells overexpressing the indicated mutants. (C) Quantitative measurement of the progesterone synthesized. Data represent the means plus SEM for three independent experiments performed at three different times. (D) Co-IP analysis with 3βHSD2 antibodies after Tom22 knockdown and expression of the indicated mutants in MA-10 cells. The blots were stained with 3βHSD2 and Tom22 antibodies independently. The bottom panel shows VDAC2 expression, showing the same amount of cells were used for each experiment. (E to I) Electron microscopy of Tom22 mutants (F to I) in ΔTom22 MA-10 cells. (E) Typical pattern of an enlarged view of mitochondrion stained with 3βHSD2 and Tom22 antibodies. (F) Expression pattern of Tom22 (bottom panel), and the top panel shows its typical localization in a mitochondrion. The small gold particles (15 nm, red arrowheads) are 3βHSD2, and the larger 55-nm particles are Tom22 (green arrows). (G) Tom22 expression is absent in ΔTom22 MA-10 cells, and the top panel shows only one mitochondrion with minimal Tom22. (F and G) Expression of Tom22 in wild-type (F) and siRNA knockdown (G) MA-10 cells. (H and I) Bottom panels show expression of N124K (H) and K141D (I) Tom22 in ΔTom22 MA-10 cells. The top panels show an enlarged mitochondrion from the bottom panel. Bars, 2.0 μm (F) and 1.0 μm (G to I). (J) RT-PCR amplification of cDNA from ΔTom22 MA-10 cells with different 3βHSD2-nested primers showing amplification of 747-bp, 394-bp, and 600-bp cDNA. The human 3βHSD2 plasmid DNA was the control for the PCR, and WT MA-10 cDNA was used as internal control for the siRNA knockdown. The positions of DNA markers (in base pairs) are shown to the right of the gel.