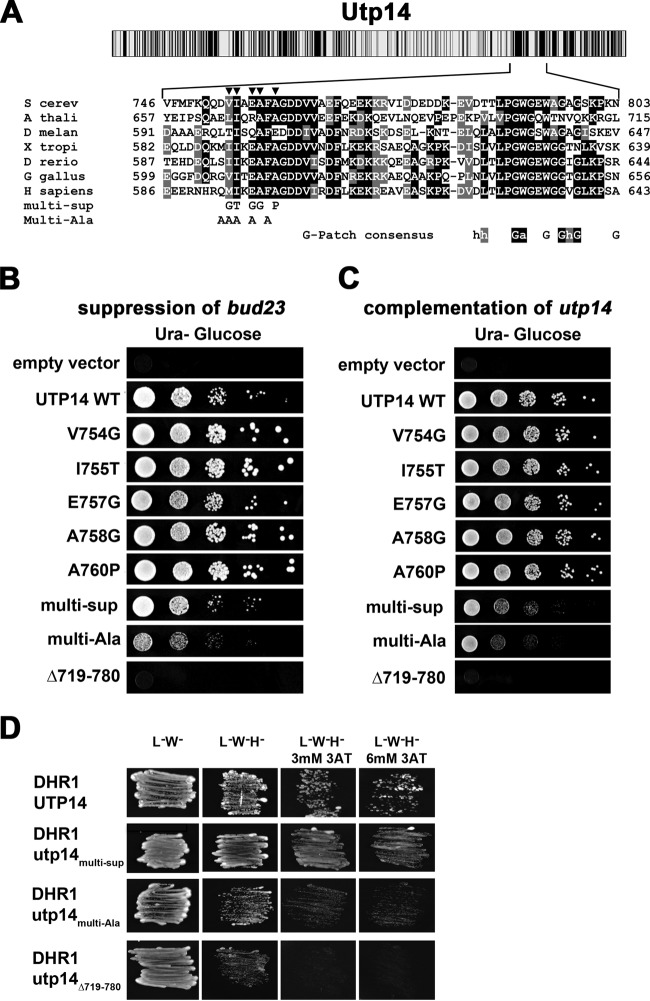
FIG 2.
Suppressing mutations map to a highly conserved motif in Utp14. (A) Cartoon showing conservation of amino acid sequence across Utp14 (top) as described in the legend to Fig. 1. Multiple-sequence alignment of the region of Utp14 containing mutations that suppress bud23Δ (bottom) was performed with T-coffee. S cerev, Saccharomyces cerevisiae; A thali, Arabidopsis thaliana; D melan, Drosophila melanogaster; X tropi, Xenopus tropicalis; D rerio, Danio rerio; G gallus, Gallus gallus; H sapiens, Homo sapiens. Positions of single amino acid substitutions that suppress bud23Δ are indicated by triangles. The amino acid changes of Utp14multi-Sup and Utp14multi-Ala are indicated. The consensus sequence of G-patch proteins is shown below the alignment. Perfect matches with invariant residues are shaded in black, and similar residues are shaded in gray. (B) WT UTP14 or the indicated mutants were expressed in bud23Δ PGAL1-UTP14 strain AJY3245, and 10-fold serial dilutions of cells were spotted onto glucose-containing medium and grown for 2 days at 30°C. (C) WT UTP14 or the indicated mutants were expressed in PGAL1-UTP14 strain AJY3243, and 10-fold serial dilutions of cells were spotted onto glucose-containing medium and grown for 2 days at 30°C. (D) Combining suppressing mutations, multiple alanine substitutions, or deletion of the region of Utp14 identified by suppressing mutations causes an increasing defect in Dhr1 interaction scored by two-hybrid assay.