FIG 6.
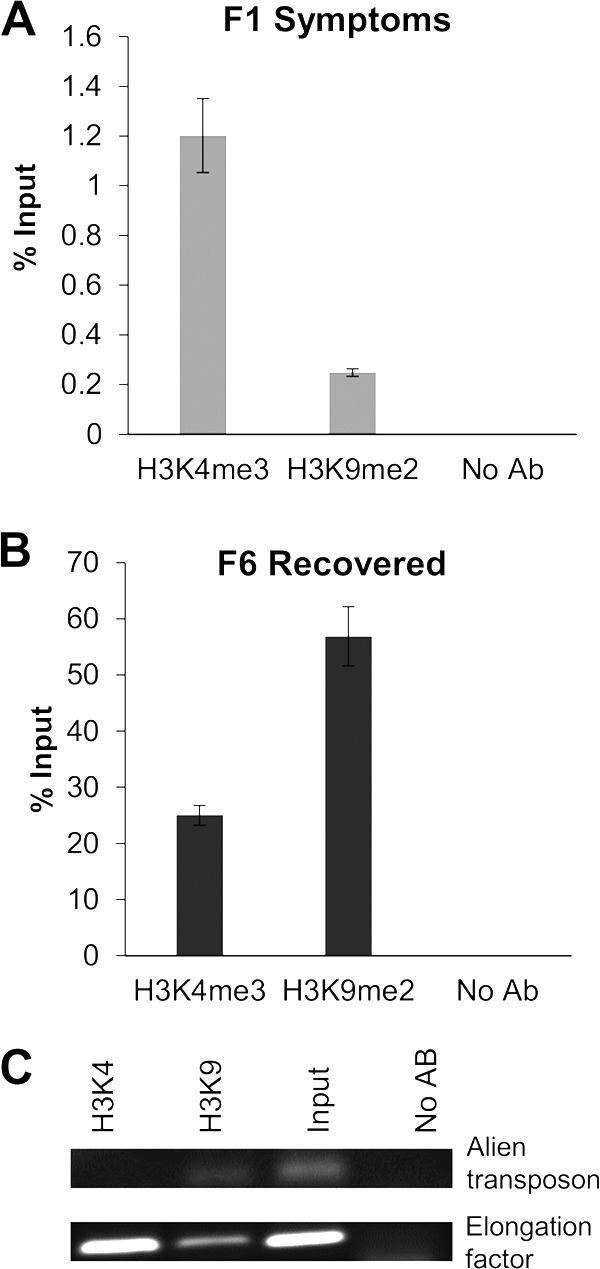
The minichromosome populations from F1S and F6R fractions present an opposite association to active and repressive chromatin histone markers. ChIP-qPCR experiments using antibodies against active (H3K4me3) and repressed (H3K9me2) chromatin markers were carried out for both minichromosome populations from F1S (A) and F6R (B) samples. Input (before immunoprecipitation, 100%) and viral pulled-down DNA were quantified by qPCR (and data are presented as percentages of the input). (C) As a control, the same antibodies (Abs) were used for ChIP assays for genes/elements known to be present in usually condensed/repressed (alien transposon) or relaxed/active (elongation factor) chromatin regions. Total DNA from an equivalent aliquot of the chromatin sample was used as the “input” control. Similarly, an aliquot of the same chromatin sample was included in the ChIP assay, but no Abs were added.
