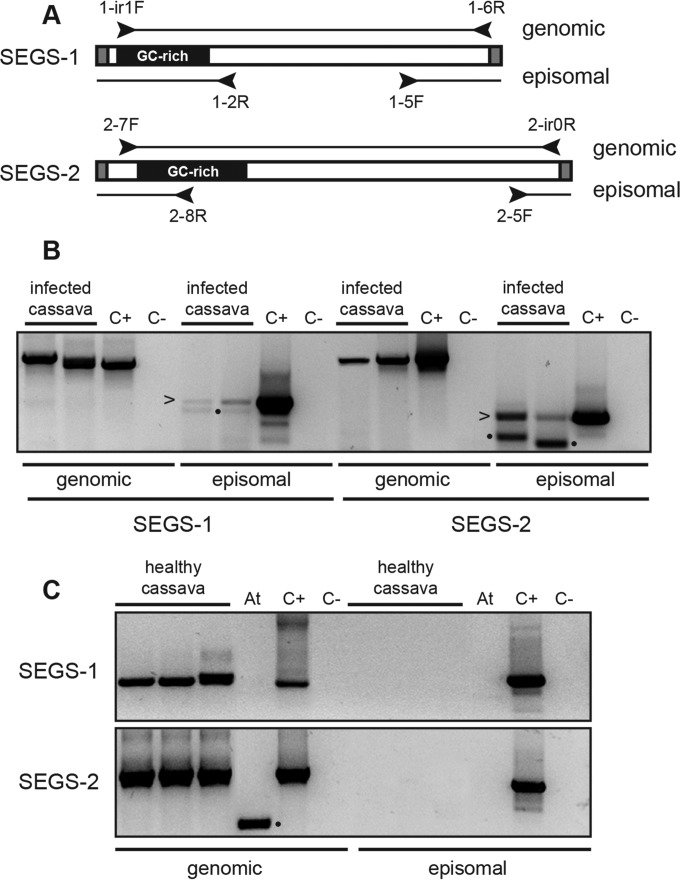
FIG 7.
Amplification of SEGS-1 and SEGS-2 episomes in infected plants. (A) The convergent primer pairs 1-hp1F/1-6R and 2-7F/2-hp-0R amplify genomic copies of SEGS-1 and SEGS-2, respectively. The divergent primer pairs 1-2R/1-5F and 2-8R/2-5F amplify circular episomal or concatemeric copies of SEGS-1 and SEGS-2, respectively. Total DNA was the template for the genomic PCR products, while RCA DNA was the template for the episomal PCR products. (B) PCR products from CMB-infected cassava samples from Cameroon. The arrowheads mark bands with sequences that match SEGS-1 or SEGS-2. (C) PCR products from healthy cassava collected from Cameroon and passaged through tissue culture. The Arabidopsis thaliana (At) DNA was a control for potential contamination during DNA isolation. C+ is the positive PCR control using a cloned DNA template. C− is the negative PCR control that lacks template DNA. Bands marked with dots are nonspecific products that were also sequenced.