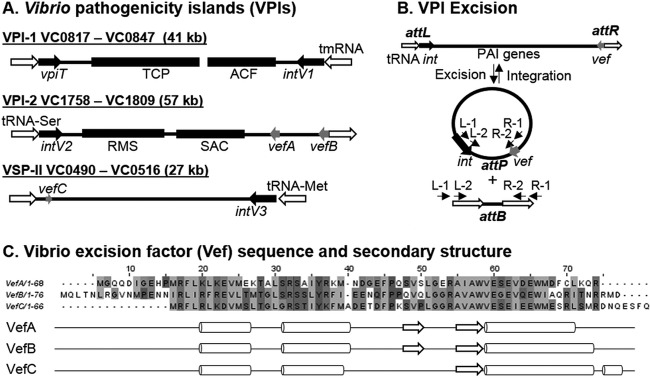
FIG 1.
Vibrio pathogenicity islands (VPIs), VPI excision, and Vibrio excision factors (Vefs). (A) VPI-1 inserts at the transfer-messenger RNA (tmRNA) gene and contains genes encoding the toxin-coregulated pilus (TCP) and accessory colonization factor (ACF) as well as an integrase (intV1) and a transposase-like gene (vpiT). VPI-2 inserts into a tRNA-serine locus and contains a restriction modification system (RMS) and a sialic acid scavenging, transport, and catabolism (SAM) region as well as an integrase (intV2) and two recombination directionality factors (RDFs) encoded by vefA and vefB. Vibrio seventh pandemic island II (VSP-II) contains a third RDF encoded by vefC. (B) VPIs can excise from the chromosome and form a circular intermediate containing an attP site and leaving a unique attB site left in the bacterial chromosome. Primers used to detect the attachment (att) sites in two-stage nested PCR assays are represented by arrows on the left and right sides of the attP and attB sites. (C) Amino acid sequence alignment of VefA, VefB, and VefC using ClustalW with default settings in JalView. A 59% identity is shared between VefA and VefC, 46% between VefA and VefB, and 49% between VefB and VefC. Predicted secondary structures of VefA, VefB, and VefC were generated using JNet Secondary Structure Prediction. Arrows represent sheets and cylinders represent helices.