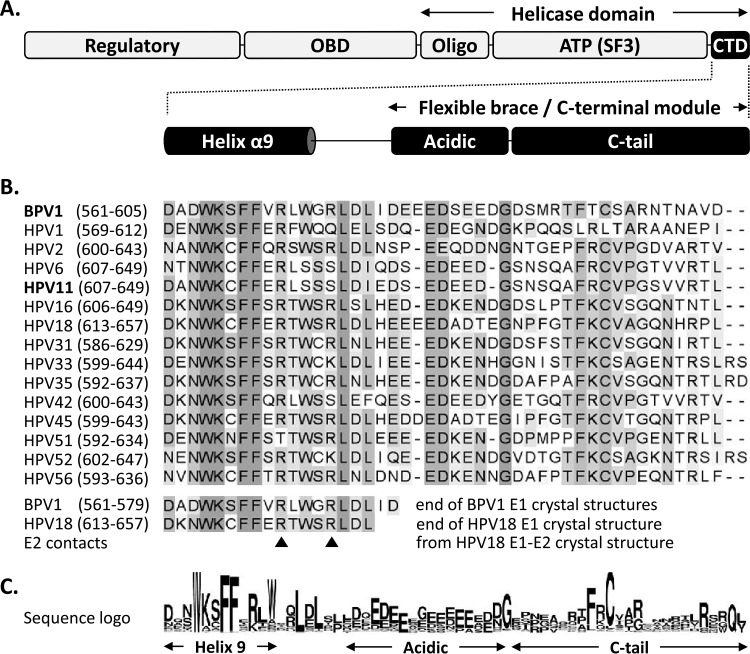
FIG 1.
Schematic representation of the papillomavirus E1 helicase highlighting the conservation of the C-terminal domain. (A) Diagram of the E1 protein showing the locations of the N-terminal regulatory region, OBD, Oligo, and ATPases associated with diverse cellular activities (AAA+) SF3 helicase/ATPase domain [ATP(SF3)] and of the ∼45-amino-acid-long CTD, which encompasses both the last alpha helix resolved in the available E1 crystal structures (helix α9) and the region previously called the flexible brace or C-terminal module, itself comprised of an acidic region (Acidic) and a C-tail subdomain separated by a linker sequence (10, 16, 17). (B) Amino acid sequence alignment of the E1 C termini from the indicated papillomavirus types (amino acid boundaries are indicated in parentheses). A large alignment of all of the E1 reference sequences deposited in the PAVE database was generated, but only those from BPV1 and 15 selected HPV types are presented for brevity. Residues are highlighted with increasingly darker shades of gray according to their degrees of conservation. The bottom of the alignment indicates the last region of the E1 C terminus that is resolved in the crystal structures of the E1 helicase domain from BPV1 (up to aa 577 and 579 in PBD structures 2V9P and 2GXA, respectively) and from HPV18 in complex with the E2 transactivation domain (PDB 1TUE) (18–20). While the major contacts between HPV18 E1 and E2 involve residues located in the ATPase domain, two potential minor contact points were identified within the C terminus (arrowheads) (20). (C) Amino acid sequence logo of the E1 C terminus generated from the alignment of all E1 reference sequences in the PAVE database (33).