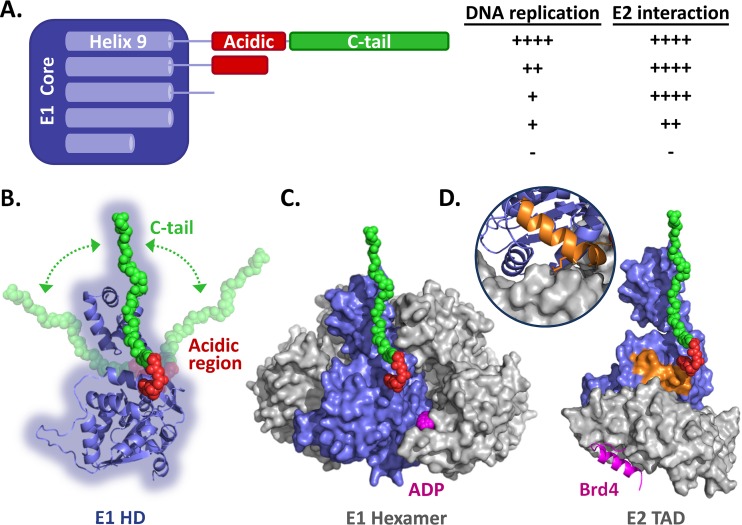
FIG 10.
Summary of the data and homology models of the E1 helicase domain. (A) Schematic representation of the HPV11 E1 core domain (aa 1 to 617) fused to the linker sequence, AR, and C-tail domain. Helix 9, which is required for the proper folding of the helicase domain, is diagrammed within the E1 core region. Also shown is a summary of the results presented here for the effects of removing the C-tail, AR, and linker sequence and of deleting part of helix 9 on the ability of HPV11 E1 to support viral DNA replication and to interact with E2 (++++, like wild-type E1; ++, ∼50% reduction; +, ∼75% reduction; −, inactive). (B) Homology model of the HPV11 E1 HD based on the structures of the analogous regions from BPV1 and HPV18 (18–20). A cartoon representation of the AR (red) and C-tail (green), drawn to scale, is shown in an extended conformation to suggest how far in space these two regions could possibly reach. The arrows are meant to represent the flexible nature of the E1 C terminus. (C) Model of the HPV11 E1 HD in its hexameric form. ADP is colored magenta. (D) Model of the HPV11 E1 HD (blue) in complex with the E2 TAD (gray) and Brd4 helix (magenta). The model is based on the structures of the HPV18 E1-E2 complex and the HPV18 E2-Brd4 complex (35). The C-terminal alpha helix of Brd4, which binds on the opposite face of the E2 TAD (gray), is colored magenta. The inset shows the interaction of helix 9 (orange) with the E2 TAD. Also shown in orange is the side chain of arginine 616 that likely contacts the E2 TAD directly, as observed for the analogous residue (arginine 622) in the HPV18 E1-E2 TAD structure (20).