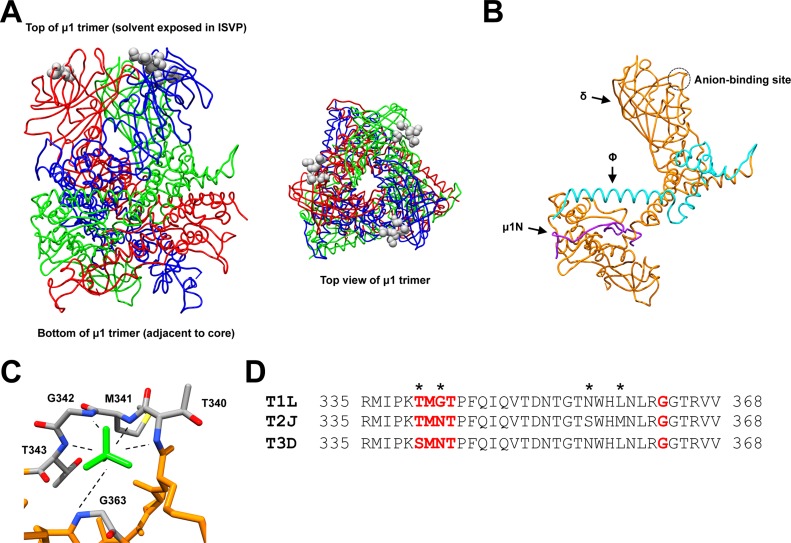
FIG 1.
Structure and sequence of the reovirus μ1 protein. (A) Side and top views (left and right, respectively) of the T1L μ1 homotrimer (52) (PDB accession number 1JMU). Individual μ1 monomers are colored in red, blue, and green. Residues corresponding to the anion-binding site are represented as gray spheres. (B) Structure of the T1L μ1 monomer (52) (PDB accession number 1JMU). Purple, μ1N; teal, Φ; orange, δ. The anion-binding site is indicated with a circle. (C) Structure of the T1L μ1 anion-binding site (52) (PDB accession number 1JMU). Residues that form the anion-binding site are labeled and colored in gray (carbon), blue (nitrogen), red (oxygen), and yellow (sulfur). The sulfate ion is colored in green. (D) Amino acid sequence alignments of the reovirus μ1 anion-binding site. Residues that correspond to the anion-binding site are bolded in red. Residues that are not conserved within the sequence region are indicated with an asterisk.T1L, reovirus type 1 Lang; T2J, reovirus type 2 Jones; T3D, reovirus type 3 Dearing.