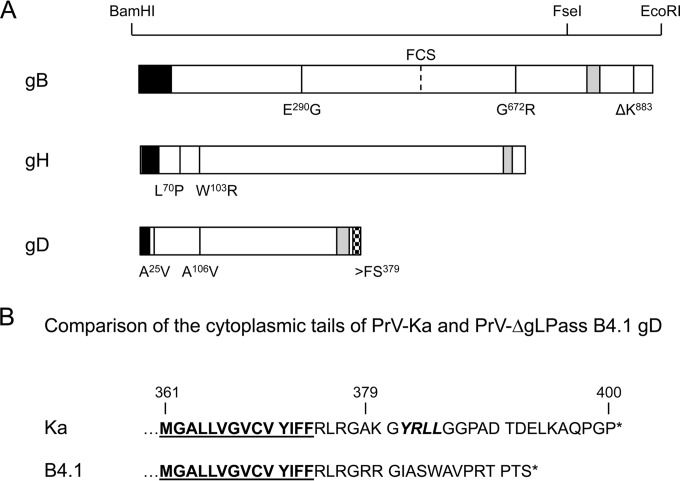
FIG 2.
Mutations in PrV-ΔgLPassB4.1. (A) Amino acid changes found in PrV-ΔgLPassB4.1 gB, gH, and gD compared to PrV-Ka are indicated at the corresponding positions in one-letter code. Restriction enzyme recognition sites used for cloning of gB mutants are indicated above the diagram. The predicted signal sequences are shown in black and the transmembrane domains in gray. The location of the furin cleavage site (FCS) in gB is given. The frameshift mutation (>FS379) in gDB4.1, which results in an altered cytoplasmic tail, is indicated by a checkered box. (B) Amino acid sequences of the gDKa and gDB4.1 C termini. Residues predicted to reside within the membrane are underlined, and the C-terminal ends are marked by asterisks. Indicated by italics is the predicted endocytosis motif in gDKa which is lost in gDB4.1 (58).