FIG 10.
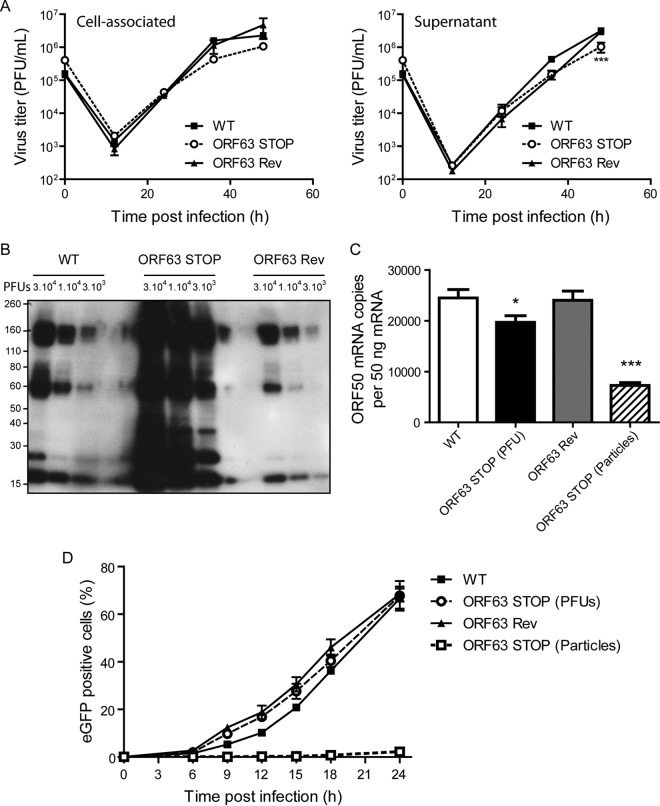
ORF63 deficiency is associated with an increased particle/PFU ratio and a deficit in entry. (A) High-MOI growth curve. BHK-21 cells were infected with WT, ORF63 STOP, and ORF63 Rev MuHV-4 strains in 6-well cluster dishes at an MOI of 1 PFU per cell. Supernatant and infected cells were harvested independently at different times after infection, and the amounts of infectious virus were determined for both kinds of samples by plaque assay on BHK-21 cells. The data are the averages from triplicate measurements ± SEMs and were analyzed by 2-way ANOVA and Bonferroni posttests. ***, P < 0.001. At time zero p.i., the inocula were retitrated to ensure that similar amounts of virus were put on the cells. (B) Comparison of the structural proteins content for different amounts of PFU (3 × 104, 1 × 104, and 3 × 103) between the different strains. MuHV-4 WT, ORF63 STOP, and ORF63 Rev stocks were compared for viral protein content by immunoblotting with anti-MuHV-4 rabbit polyserum. (C) A total of 106 BHK-21 cells were infected with WT, ORF63 STOP, and ORF63 Rev MuHV-4 strains at an MOI of 0.5 PFU/cell. For the ORF63 STOP strain, an additional sample of cells infected by a number of particles equivalent to the WT and ORF63 Rev strains was added. Six hours later, RNA was extracted, reverse transcribed, and assayed for ORF50 expression by qPCR amplification. The data are averages from triplicate measurements ± SEMs and were analyzed by 1-way ANOVA and Bonferroni posttests. (D) BHK-21 cells were exposed to eGFP expressing (BAC+) WT, ORF63 STOP, and ORF63 Rev strains (0.5 PFU/cell). For the ORF63 STOP strain, an additional sample of cells infected by a number of particles equivalent to those of the WT and ORF63 Rev strains (determined by Western blotting) was added. After binding for the times indicated, the cells were washed with PBS and assayed by flow cytometry for eGFP expression. The data are the averages ± SEMs from triplicate measurements. The data were analyzed by 2-way ANOVA and Bonferroni posttests. ***, P < 0.001.