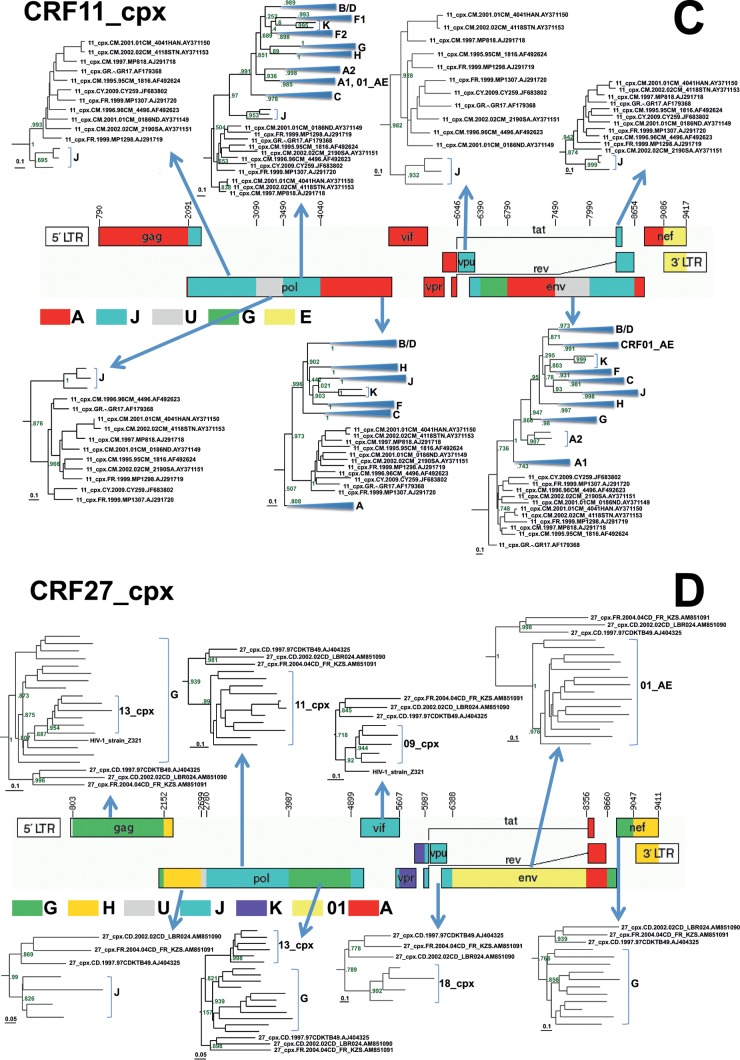
FIG 2.
Reanalysis of the origins of recombinationally derived sequence fragments within some CRFs from the Congo basin that have previously been described in the literature. Maximum-likelihood trees indicating the phylogenetic relationships between segments of recombinationally derived sequence from these CRFs and different HIV-1M subtypes are shown. ML trees were constructed from aligned sequences corresponding to these segments using FastTree 2 (23). Numbers at the tree branches indicate support for these branches using the Shimodaira-Hasegawa-like branch support test. For the sake of clarity, only subtrees that included the divergent, difficult-to-classify sequences were included, except in panel A, where all the subtrees are shown. The graphical genome maps of the CRFs analyzed here were obtained from the Los Alamos HIV database (21) and represent the currently accepted breakpoint and parental subtype annotations for these CRFs.