FIG 2.
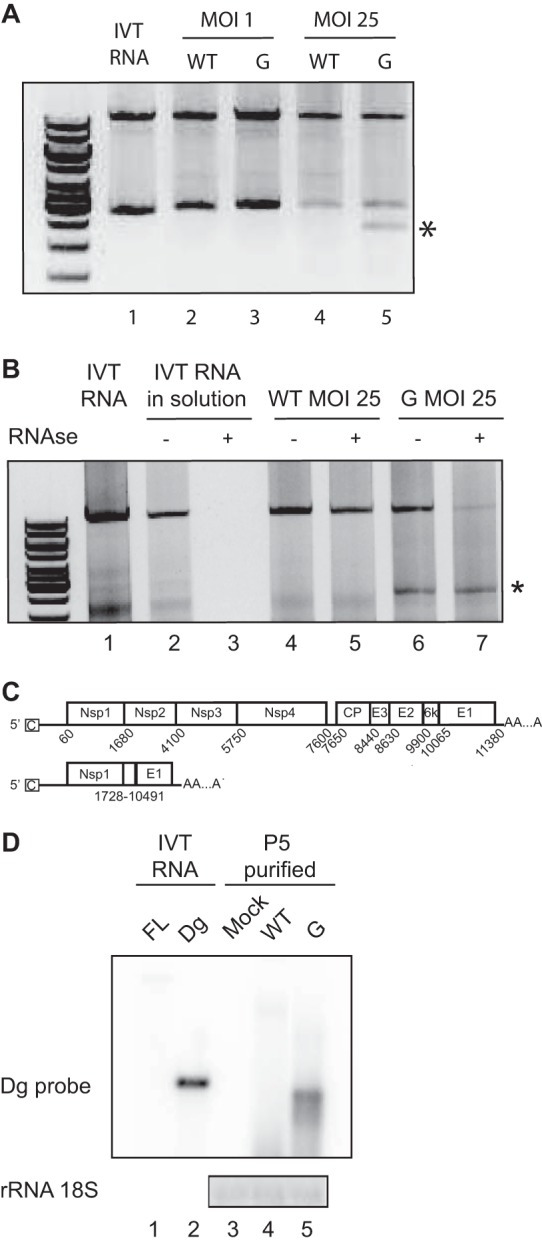
Defectives particles can be detected during SINV-G infection. (A) Detection of SINV-WT (WT) and SINV-G (G) genomic sequences by RT-PCR after 24 h of infection at an MOI of 1 or 25. In vitro-transcribed RNA (lane 1) was used as a control. The top band corresponds to the full-length genome. An extra low-molecular-weight band is indicated by an asterisk. (B) Viral supernatants from a 24-h infection with SINV-WT (lanes 4 and 5) or SINV-G (lanes 6 and 7) were either untreated or treated with RNase A (−, absence; +, presence) and then subjected to RT-PCR as described for panel A. In vitro-transcribed RNA (lane 1) was used as an RT-PCR control. IVT RNAs incubated in solution (lane 2) and treated with RNase A (lane 3) were used as RNase controls. (C) Schematic of SINV full-length genome and the defective genome, Dg. Schematic shows the 5′ and 3′ untranslated regions with cap (C) and poly(A) tail and the intergenic region containing the subgenomic promoter (arrow) for structural proteins. Each protein-coding gene is depicted as a separate box, with positions of start and stop nucleotides numbered below. The deleted nucleotides are numbered below the Dg scheme. (D) Five undiluted passages were performed on BHK-21 cells with SINV-WT (lane 4) or SINV-G (lane 5) virus, or cells were mock infected (lane 3). Progeny virions were treated with RNase and purified by ultracentrifugation. Viral RNA was probed by Northern blotting using a probe specifically targeting the Dg breakpoint. In vitro-transcribed (IVT) RNAs corresponding to full-length (FL) and Dg genomes, not treated with RNase, were used as positive controls for detection (lanes 1 and 2).
