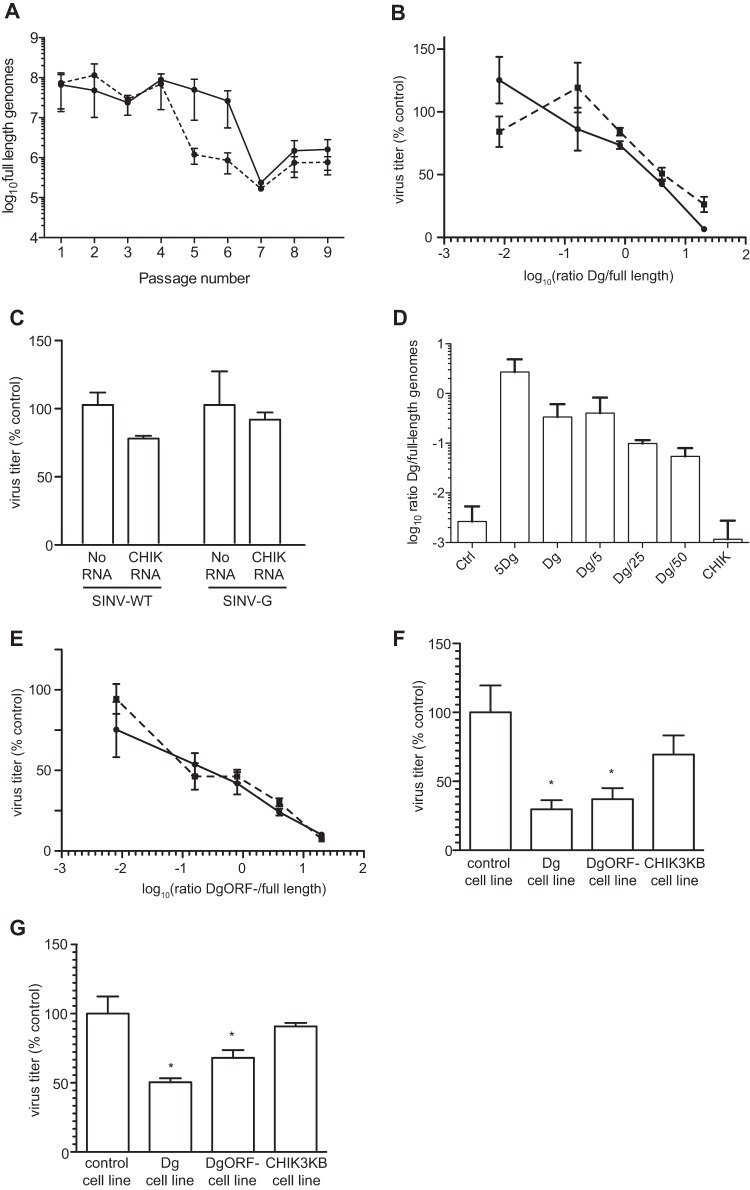
FIG 4.
SINV defective genomes interfere with full-length virus replication. (A) The number of full-length genomes in high-MOI passages of the SINV-WT (solid line) and SINV-G (dashed line) was assessed by qRT-PCR. Means and standard errors of the means are shown (n = 3). (B) 293T cells were transfected with a fixed amount of full-length SINV-WT (solid line) or SINV-G (dashed line) RNA and an increasing amount of Dg RNA. Titer was measured by plaque assay at 24 h posttransfection. Means and standard errors of the means are shown (n = 3). (C) Chikungunya virus replication-defective RNA (CHIK RNA) does not significantly interfere with SINV-WT or SINV-G virus replication compared to replication in infected cells with no additional RNA added (No RNA). Means and standard errors of the means are shown (n = 3; no significant differences were detected by a two-tailed student t test). (D) Viral supernatant from the transfection presented in panel B was subjected to RNase A treatment before qRT-PCR to measure the Dg/full-length genome ratio as previously described. The x axis indicates the ratio of Dg/full-length RNA initially transfected. Means and standard errors of the means are shown (n = 3). (E) 293T cells were transfected with a fixed amount of full-length SINV-WT (solid line) or SINV-G (dashed line) RNA and an increasing amount of Dg RNA mutated in its nsP1 starting codon (DgORF−). Titer was measured by plaque assay at 24 h posttransfection. Means and standard errors of the means are shown (n = 3). (F and G) Stable cell lines expressing Dg, DgORF−, or CHIK3KB were infected with either SINV-WT (F) or SINV-G (G), and titers were measured at 16 h postinfection. Means and standard errors of the means are shown (n = 3; *, P < 0.05, for sample values compared to those of the wild-type, by one-way analysis of variance with a Bonferroni posttest).